MicroRNAs in atopic dermatitis: a review
Abstract
Atopic dermatitis (AD) is a chronic and relapsing inflammatory skin disease, generally the first clinical manifestation of atopy and the start of atopic march. Effective treatment of AD could potentially interrupt the progression of atopic march. MicroRNAs (miRNAs), a recently described class of gene expression regulators in inflammatory conditions, affect expression of numerous proteins. The role of miRNAs has been investigated in several atopic conditions, including asthma, eosinophilic esophagitis, allergic rhinitis as well as atopic dermatitis. They have been shown to be involved in the morphogenesis of skin. The therapeutic effects of inhibition or overexpression of miRNAs have been demonstrated in murine models. Considering their role as master switches of complex cellular processes, they could be potential therapeutic targets for inflammatory skin conditions including atopic dermatitis. MiRNAs can be detected in different cell-free body fluids, such as serum, plasma, urine and saliva, raising the obvious question whether they can be used as biomarkers of disease. This review article summarizes what is known so far abut miRNAs and atopic dermatitis.
Keywords
Introduction
Atopic dermatitis (AD) is a chronic and relapsing inflammatory skin disease. It is generally the first clinical manifestation of atopy and the start of atopic march[1,2]. Atopic march refers to the phenomenon of progression of atopic dermatitis to asthma and allergic rhinitis[3,4]. The defective skin barrier in patients with AD might allow allergen exposure through the epidermis initiating systemic allergen sensitization leading to development of food allergy and asthma[4]. Therefore, effective treatment of AD could potentially interrupt the progression of atopic march. A deeper understanding of the pathogenesis of this condition could provide tools for early intervention. The pathogenesis of AD is complex, involving genetic predisposition with a strong influence of innate and adaptive immune responses, as well as environmental factors[5]. Interplay between genetic and environmental factors leading to skin barrier dysfunction drives the natural history of the disease[5]. Specifically, Th2 and Th22 pathways, along with inflammatory cytokines and mediators are implicated[6].
Inflammation in atopic dermatitis is biphasic: an initial Th2 phase precedes a chronic phase in which Th0 cells and Th1 cells predominate[7]. The Th2 cytokines interleukin (IL)-4, IL-5 and IL-13 predominate in acute lesions, and in chronic lesions, there is increase of interferon-γ (IFN-γ) and IL-12[7]. There is a complex interaction between the keratinocytes and T cell subpopulations, and their respective cytokine milieu. There are numerous T cells in the skin, approximately 106 memory T cells per square cm of body surface area. Keratinocytes in eczema skin produce high levels of thymic stromal lymphopoietin (TSLP) that signals dendritic cells to drive Th2 polarization[7]. Th17 T cells are a subset of memory CD4+ T cells that produce IL-17A and IL-17F[8]. Higher Th17 cell count has been shown to be crucial to development of initial Th2-related phase, while reduced levels are seen in the chronic Th-1 mediated phase[9]. IL-17 is a proinflammatory cytokine highly expressed in acute AD, but barely detectable in chronic AD. It has been described as a master regulator of antimicrobial peptides (AMPs)[10]. Low levels of IL-17 in the chronic phase of AD correlate with higher risk of cutaneous super infections[11]. Th22 cells have been characterized recently as IL-22-producung T cells[12]. IL-22 which is the prototypical Th22 cytokine is highly expressed in AD. High levels of chemokine ligand (CCL) 27 can be found in chronic AD lesions[13]. This chemokine has been shown to induce homing of Th22 cells in lesional AD skin. IL-22 is a member of the IL-10 cytokine family. It also induces AMP production, although less efficiently that IL-17. It is responsible for epidermal hyperplasia and downregulates filaggrin expression[14].
The “outside-in” hypothesis proposes that the defective barrier allows penetration of allergens and microbes into the skin of patients with AD[5]. At the opposite end of the debate, the inside-out hypothesis proposes a polarized immune response being the cause of a defective skin barrier[5]. A strong association has been established between loss-of-function mutations in skin barrier gene encoding filaggrin (FLG) and AD[15,16]. Up to a 40% of patients with AD have FLG mutations[17]. The barrier defect in AD patients with FLG mutations promotes enhanced allergen penetration leading to increased production of TSLP by keratinocytes. This in turn leads to a Th2-type milieu. In fact, TSLP has been called a “master switch for allergic inflammation”. Aside from FLG, loss-of-function mutations in serine protease inhibitors (e.g., SPINK5) are associated with epidermal barrier dysfunction through increased Th1 responses[18]. Innate pattern recognition receptors such as Toll-like receptors are expressed on keratinocytes and antigen-presenting cells in the skin[19]. The TLRs, when stimulated by tissue injury and microbe invasion, release AMPs which in turn limit penetration of microbes and allergens. Keratinocytes from skin of patients with AD may not produce enough AMPs to control Staphylococcus aureus and viral replication, predisposing these patients to S. aureus and viral infections. The Th2, Th22 and TH17 cytokine pathways are associated with the acute lesions of AD[6]. In patients with chronic disease, there is intensification of these pathways along with appearance of a significant Th1 component[6].
A recently described class of gene expression regulators in inflammatory conditions is short single-stranded RNA molecules termed microRNAs (miRNAs)[20]. MiRNAs, which are 19-25 nucleotides in length, cause gene silencing through degradation of target RNAs or inhibition of translation. Since each miRNA can have hundreds of target genes, together miRNAs can affect expression of numerous proteins[20]. It is estimated that miRNAs collectively influence approximately 30% of genes[21]. The importance of miRNAs is evidenced by the fact that deletion of Dicer enzyme, required for maturation of precursor miRNA is lethal in mice[22]. They have been shown to be key regulators in early development, proliferation, stress response and apoptosis[23]. They have been implicated in the pathogenesis of cancer, developmental abnormalities and cardiovascular diseases[24]. Not only does each miRNA target hundreds of genes, the individual genes are targeted by several miRNAs adding to the complexity of these interactions. MiRNAs are, therefore, well positioned to affect allergic inflammatory pathways. Their role has been investigated in asthma, eosinophilic esophagitis, allergic rhinitis as well as atopic dermatitis. They have been implicated in the morphogenesis of murine skin[25]. Murine disease models have shown the therapeutic effects of inhibition or overexpression of miRNAs, thus making them potential therapeutic and diagnostic candidates.
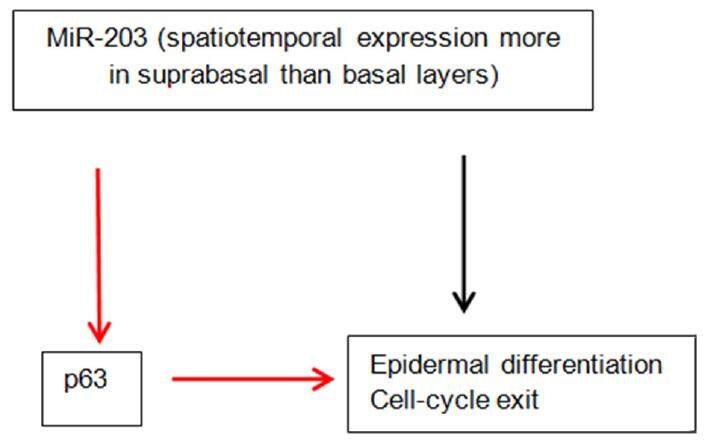
The importance of miRNAs in skin development is underscored by the fact that their global ablation by targeting Dicer 1 prevents hair follicles from invaginating, disrupting the epidermal morphology[25,26]. MiR-203 has been identified as a skin and keratinocyte specific miRNA. Its expression is higher in the suprabasal layers compared to the basal layer[26]. It can induce keratinocyte differentiation. Its critical role in epidermal morphogenesis is supported by the observation that overexpression in mice results in a thinner epidermis and decreased proliferation[27]. MiR-203 directly targets p63 which is critical to the development of epidermis[28]. p63 is an essential regulator of stem cell maintenance in stratified epithelial cells. Wei et al found that p63 and miR-203 have mutually exclusive expression pattern within the epidermis during human skin development[29]. MiR-203 directly represses the expression of p63 [Figure 1]. It has been suggested that miR-203 defines a molecular boundary between the proliferative basal layer and differentiating suprabasal layers. Premature expression of miR-203 can cause repression of p63[27]. Moreover, miR-203 affects the post-translational expression of suppressor of cytokine signaling-3 (SOCS-3), which is a negative regulator of Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway[26]. There is compelling evidence that it induces cell-cycle exit and represses “stemness” in epidermal progenitors. In the following review, we have summarized what is known so far about miRNAs and AD.
MiR-155 is overexpressed in skin of patients with AD
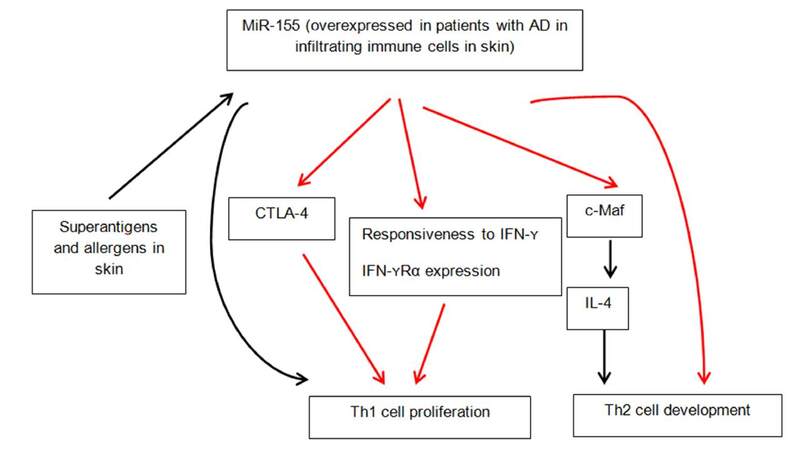
MiR-155 represents a typical multifunctional miRNA with crucial role in a variety of physiological and pathological processes such as hematopoietic lineage differentiation, immunity, inflammation, cancer and cardiovascular diseases[30]. It was the first microRNA to be studied in miR-deficient mice, establishing its pleiotropic effects on dendritic, T cells and B cells. Multiple studies have established a critical role of miR-155 in T cell differentiation. It has been shown to indirectly promote Th1 polarization. It has been suggested that miR-155 promotes Th1 only in the absence of Th2 polarizing cytokines[31]. It targets c-Maf, a transactivator of IL-4 promoter to attenuate Th2 pathway[32]. Dicer is a nuclease essential to miR biogenesis, the deficiency of which in CD4+ T cells leads to diminished proliferation and increased cell death upon activation[33]. CD4+ T cells deficient in Dicer are predisposed to differentiation into IFN-γ producing Th1 cells. MiR-155 is the only miR known to influence CD4+ T cell differentiation, via known targets SOCS-1 and c-Maf. Banerjee et al.[33] showed that miR-155 over-expression favored CD4+ T cell differentiation towards Th1 pathway while its antagonism biased the cells towards Th2 differentiation.
Global miRNA expression in healthy and lesional skin of patients with atopic dermatitis was compared by using TaqMan microRNA low density arrays by Sonkoly et al.[34]. They identified 44 miRNAs that were significantly differentially expressed in AD lesions. Of these, 34 were downregulated and 10 were upregulated compared to healthy skin. The miRNA with the highest overexpression was miR-155. MiR-155 is known to be involved in carcinogenesis, immune cell development and regulation of innate and adaptive immune responses[35]. Further analysis of healthy human organs and tissues revealed high levels of expression of miR-155 in thymus, spleen and lung (organs rich in immune cells). Healthy skin had relatively low levels of expression of miR-155 compared with other organs. Further analysis of a panel of cells present in healthy skin, inflamed skin or both revealed expression in T cells, dendritic cells, fibroblasts and mast cells, which have all been implicated in the pathogenesis of chronic skin inflammation. Immunohistochemical staining and phenotyping established that CD4+ cell population was the largest in both atopic dermatitis and control skin, followed by dendritic cells. It was concluded that CD4+ T cells (TH cells) were major contributors to the increased expression of miR-155 in patients with AD[34]. Furthermore, miR-155 was found to be involved in TH cell activation and differentiation. Moreover, quantitative real-time PCR analyses showed that miR-155 mRNA was significantly induced after exposure to staphylococcal superantigen. Staphylococcus aureus colonizes the skin of 90% of patients with AD and represents a trigger factor for skin inflammation. Dust mite exposure via atopy patch testing with dust mite allergen also led to increased miR-155 levels after 48 h of allergen exposure[34]. Furthermore, CTLA-4, a major inhibitory molecule of T-cell responses was identified as a novel target of miR-155, demonstrating that miR-155 regulates the proliferation of TH cells. Blockade of CTLA-4 has been shown to enhance allergic sensitization and eosinophilic airway inflammation in genetically predisposed mice with increased eosinophil numbers, IgE levels and IL-13 levels[36]. The successful treatment of chronic inflammatory diseases by the CTLA-4-immunoglobulin fusion protein abatacept is evidence of its ability to efficiently inhibit inflammatory responses[37]. MiR-155-mediated suppression of CTLA-4 expression appears to be crucial in the pathogenesis of AD, making miR-155 a promising target for therapeutic intervention. The multifunctional role of miR-155 is summarized in Figure 2.
MiR-146a expression is increased in keratinocytes in AD
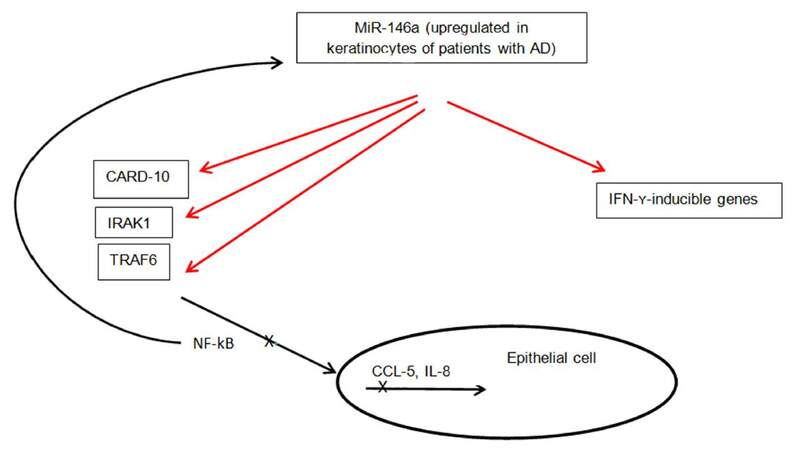
Another microRNA that has been shown to play a key role in the inflammatory process of AD is miR-146a. Leukocyte-rich organs such as thymus and spleen had high expression of miR-146a, as opposed to healthy skin, indicating the infiltrating leukocytes in eczematous and psoriatic skin express mi-R146a[38]. Skin inflammation in AD is complicated with secondary infections, such as Staphylococcus aureus that lead to activation of the nuclear factor kappa B (NF-kB) pathway in keratinocytes[19,39]. The expression of miR-146a is dependent on NF-kB. It moreover, suppresses the NF-kB signaling pathway through targeting of IL-1 receptor-associated kinase 1 (IRAK1), TNF receptor-associated factor 6, v-rel avian reticuloendotheliosis viral oncogene homolog B (RELB), and caspase recruitment domain-containing protein 10 (CARD10)[38,40-43]. MiRNA-146a may, therefore, be involved in a negative feedback mechanism of TNF-α signaling pathway. Mice deficient in miR-146a have been shown to spontaneously develop autoimmunity due to activation of NF-kB[43,44]. Rebane et al.[45] showed that the level of miR-146a is increased in keratinocytes from patients with AD. MiR-146a upregulation was associated with inhibition of numerous proinflammatory factors, including IFN-γ-inducible genes. IFN-γ induces expression changes of thousands of genes in keratinocytes[14]. MiR-146a was found to downregulate 410 genes in IFN-γ-stimulated keratinocytes pointing to a strong anti-inflammatory effect of this microRNA in chronic AD in this study[45]. Enhanced expression of NF-kB-dependent miR-146a in lesional and nonlesional skin of patients with AD has been demonstrated as well[38]. Its expression is increased in skin of patients with psoriasis also supporting the assumption that this is a general characteristic of inflammation in skin[26]. The anti-inflammatory function of miR-146a in the skin is thought to involve targeting of NF-kB pathway activating elements CARD10 and IRAK1, and suppression of IFN-γ-inducible genes CCL5, CCL8 and ubiquitin D (UBD)[45]. CARD10 and UBD could be novel factors linking the IFN-γ and NF-kB pathways. CCL5 and CCL8 are known to attract T cells, macrophages and eosinophils[46,47]. Figure 3 summarizes the above information on role of miR-146a in skin inflammation. Considering the important role of miR-146a in suppression of innate immune responses in keratinocytes, its overexpression may have a potential role in treatment of AD and other inflammatory skin conditions. The success of TNFα blockers in treatment of psoriasis leads us to believe that miR-146a could be a novel therapeutic target as well.
MiRNAs in serum and urine
MiRNAs have been found in serum and plasma, urine, tears and amniotic fluid making them promising biomarkers for various diseases[48,49]. Lv et al.[50] performed a genome-wide miRNA profiling in serum and urine of children with AD. The study confirmed stable expression of miRNAs in serum and urine. Ten serum miRNAs and 17 urinary miRNAs were identified as significantly differentially expressed between children with AD and controls. MiR-483-5p and miR-205 were upregulated in both serum and urine in children with AD compared to controls. Patients with higher IgE levels had significantly higher expression of miR-203 and miR-483-5p in serum compared to controls. MiR-483-5p in serum was found to be significantly associated with other atopic conditions such as rhinitis and/or asthma, reflecting the multi-organ/tissue involvement of atopic conditions. MiR-203 was upregulated in serum but was markedly decreased in urine of children with AD[50]. Patients with higher IgE levels in serum had significantly lower expression of miR-203 in urine, but higher expression in serum compared to controls. This contrary phenotype of miR-203 could due to its expression from other organs such as the esophagus. MiR-203 has previously been identified as a keratinocyte-specific miRNA overexpressed in psoriasis[26]. Serum levels of miR-205 have been found to be significantly lower among patients with lupus compared to controls, and are thought to positively correlate with renal function[49]. It was, moreover, demonstrated that upregulated miR-203 in serum was significantly associated with expression of sTNFRI and sTNFRII. However, neither sTNFRI nor sTNFRII were shown to be the direct downstream targets of miR-203.
Sonkoly et al.[26] have also described a specific miRNA expression profile for psoriasis. They performed a comprehensive analysis of all human miRNAs registered in mirBase 8.0 in skin lesions of patients with psoriasis and compared it to expression in healthy skin and lesional skin of patients with AD. Increased psoriasis-specific overexpression of miR-203 and miR-146 was detected. MiR-21 was significantly upregulated in both psoriasis and AD, compared to healthy skin. A target of miR-203 is suppressor of cytokine signaling (SOCS-3) which has a role in inflammatory responses and keratinocyte functions. The upregulation of miR-203 was found to be associated with down-regulation of SOCS-3[26]. On the contrary, the expression of miR-125b was decreased significantly in both psoriasis and AD[26]. Another interesting observation in that study was that miR-203 was expressed more than 100-fold higher in skin compared to other organs, including esophagus and cervix that contain squamous epithelium. However, mature forms of miR-146a, miR-21 and miR-125b were detectable in all studied organs. A synopsis of the major miRNA players in atopic dermatitis is provided in Table 1.
MicroRNA expression and functions in atopic dermatitis
| MicroRNA | Expression and functions |
|---|---|
| miR-155 | One of the most upregulated microRNAs in AD lesions, primarily expressed in infiltrating TH cells, downregulates CTLA-4, enhances TH cell proliferation, promotes TH1 differentiation |
| miR-146a | Increased expression in keratinocytes and chronic lesional skin in AD, inhibits expression of proinflammatory factors including IFN-γ-inducible genes and AD-associated genes CCL5, CCL8 and UBD, suppresses NF-κB signaling pathway by targeting IRAK1, TRAF6, RELB, CARD10, decreases allergic skin inflammation |
| miR-203 | Keratinocyte-specific, promotes epidermal differentiation, targets p63 which is a regulator of stem-cell maintenance, increased levels in serum, decreased levels in urine of children with AD |
| miR-483-5p | Increased levels in serum of children with AD, associated with rhinitis and/or asthma |
Since their discovery in mammalian cells approximately 10 years ago, the role of miRNAs in immune system has been extensively studied. MiRNAs can be detected in different cell-free body fluids, such as serum, plasma, urine and saliva. This obviously raises the question whether serum miRNAs can be used as biomarkers of disease. Numerous studies have shown that miRNAs can be used as diagnostic as well as prognostic markers in various cancers, tissue injury and inflammation. The connection between miRNAs and allergic inflammatory conditions such as asthma, allergic rhinitis and eosinophilic esophagitis is being extensively investigated. Only a few studies have delved into the role of miRNAs in atopic dermatitis. While we are learning about the expression profile of miRNAs in inflammatory skin conditions and their impact on inflammation, the question that needs to be answered is what drives this aberrant expression. Considering their role as master switches of complex cellular processes, they could be potential therapeutic targets for inflammatory skin conditions including atopic dermatitis. A potential challenge with biological treatments is the inbuilt redundancy in signaling pathways which allows natural compensation of effects of biologics targeting single genes or proteins. MiRNAs, however, target multiple genes simultaneously, hence, may offer an opportunity to modulate the functioning of whole pathways rather than individual components. With accumulating knowledge about the role of miRNAs in inflammation, along with development of safe, effective and specific methods of miRNA delivery systems, these novel gene expression regulators could potentially be used as personalized therapeutics in the near future.
Declarations
Authors’ contributionsN. Bhardwaj contributed solely to the paper.
Financial support and sponsorshipNone.
Conflicts of interestThere are no conflicts of interest.
Patient consentNot applicable.
Ethics approvalNot applicable.
Copyright© The Author(s) 2017.
REFERENCES
1. Schmitt J, Langan S, Deckert S, Svensson A, von Kobyletzki L, Thomas K, Spuls P; Harmonising Outcome Measures for Atopic Dermatitis (HOME) Initiative. Assessment of clinical signs of atopic dermatitis: a systematic review and recommendation. J Allergy Clin Immunol 2013;132:1337-47.
2. Shaw TE, Currie GP, Koudelka CW, Simpson EL. Eczema prevalence in the United States: data from the 2003 National Survey of Children's Health. J Invest Dermatol 2011;131:67-73.
3. Spergel JM. From atopic dermatitis to asthma: the atopic march. Ann Allergy Asthma Immunol 2010;105:99-106; quiz 107-9, 117.
4. Spergel JM, Paller AS. Atopic dermatitis and the atopic march. J Allergy Clin Immunol 2003;112:S118-27.
5. Leung DY, Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment approaches. J Allergy Clin Immunol 2014;134:769-79.
6. Gittler JK, Shemer A, Suárez-Fari-as M, Fuentes-Duculan J, Gulewicz KJ, Wang CQ, Mitsui H, Cardinale I, de Guzman Strong C, Krueger JG, Guttman-Yassky E. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol 2012;130:1344-54.
8. Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol 2005;6:1133-41.
9. Koga C, Kabashima K, Shiraishi N, Kobayashi M, Tokura Y. Possible pathogenic role of Th17 cells for atopic dermatitis. J Invest Dermatol 2008;128:2625-30.
10. Peric M, Koglin S, Dombrowski Y, Gross K, Bradac E, Büchau A, Steinmeyer A, Zügel U, Ruzicka T, Schauber J. Vitamin D analogs differentially control antimicrobial peptide/"alarmin" expression in psoriasis. PLoS One 2009;4:e6340.
11. Toda M, Leung DY, Molet S, Boguniewicz M, Taha R, Christodoulopoulos P, Fukuda T, Elias JA, Hamid QA. Polarized in vivo expression of IL-11 and IL-17 between acute and chronic skin lesions. J Allergy Clin Immunol 2003;111:875-81.
12. Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol 2009;10:864-71.
13. Nograles KE, Zaba LC, Shemer A, Fuentes-Duculan J, Cardinale I, Kikuchi T, Ramon M, Bergman R, Krueger JG, Guttman-Yassky E. IL-22-producing "T22" T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol 2009;123:1244-52.e2.
14. Nograles KE, Zaba LC, Guttman-Yassky E, Fuentes-Duculan J, Suárez-Fari-as M, Cardinale I, Khatcherian A, Gonzalez J, Pierson KC, White TR, Pensabene C, Coats I, Novitskaya I, Lowes MA, Krueger JG. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol 2008;159:1092-102.
15. Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med 2011;365:1315-27.
16. McAleer MA, Irvine AD. The multifunctional role of filaggrin in allergic skin disease. J Allergy Clin Immunol 2013;131:280-91.
17. Mohiuddin MS, Ramamoorthy P, Reynolds PR, Curran-Everett D, Leung DY. Increased compound heterozygous filaggrin mutations in severe atopic dermatitis in the United States. J Allergy Clin Immunol Pract 2013;1:534-6.
18. Samuelov L, Sprecher E. Peeling off the genetics of atopic dermatitis-like congenital disorders. J Allergy Clin Immunol 2014;134:808-15.
19. Kuo IH, Yoshida T, De Benedetto A, Beck LA. The cutaneous innate immune response in patients with atopic dermatitis. J Allergy Clin Immunol 2013;131:266-78.
20. Rebane A, Akdis CA. MicroRNAs: Essential players in the regulation of inflammation. J Allergy Clin Immunol 2013;132:15-26.
21. Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science 2008;319:1789-90.
22. Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs: novel regulators in skin inflammation. Clin Exp Dermatol 2008;33:312-5.
23. Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol 2013;132:3-13; quiz 4.
25. Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 2006;38:356-62.
26. Sonkoly E, Wei T, Janson PC, Sääf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Ståhle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2007;2:e610.
27. Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing 'stemness'. Nature 2008;452:225-9.
28. Candi E, Cipollone R, Rivetti di Val Cervo P, Gonfloni S, Melino G, Knight R. p63 in epithelial development. Cell Mol Life Sci 2008;65:3126-33.
29. Wei T, Orfanidis K, Xu N, Janson P, Ståhle M, Pivarcsi A, Sonkoly E. The expression of microRNA-203 during human skin morphogenesis. Exp Dermatol 2010;19:854-6.
30. Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 2009;1792:497-505.
31. Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science 2007;316:604-8.
32. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science 2007;316:608-11.
33. Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol 2010;40:225-31.
34. Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, Stenvang J, Kauppinen S, Alenius H, Lauerma A, Homey B, Winqvist O, Ståhle M, Pivarcsi A. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol 2010;126:581-9.e1-20.
35. Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J Cell Mol Med 2009;13:24-38.
36. Hellings PW, Vandenberghe P, Kasran A, Coorevits L, Overbergh L, Mathieu C, Ceuppens JL. Blockade of CTLA-4 enhances allergic sensitization and eosinophilic airway inflammation in genetically predisposed mice. Eur J Immunol 2002;32:585-94.
37. Abrams JR, Lebwohl MG, Guzzo CA, Jegasothy BV, Goldfarb MT, Goffe BS, Menter A, Lowe NJ, Krueger G, Brown MJ, Weiner RS, Birkhofer MJ, Warner GL, Berry KK, Linsley PS, Krueger JG, Ochs HD, Kelley SL, Kang S. CTLA4Ig-mediated blockade of T-cell costimulation in patients with psoriasis vulgaris. J Clin Invest 1999;103:1243-52.
38. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 2006;103:12481-6.
39. Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part II: immune cell subsets and therapeutic concepts. J Allergy Clin Immunol 2011;127:1420-32.
40. Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-κB. Immunol Rev 2012;246:205-20.
41. Etzrodt M, Cortez-Retamozo V, Newton A, Zhao J, Ng A, Wildgruber M, Romero P, Wurdinger T, Xavier R, Geissmann F, Meylan E, Nahrendorf M, Swirski FK, Baltimore D, Weissleder R, Pittet MJ. Regulation of monocyte functional heterogeneity by miR-146a and Relb. Cell Rep 2012;1:317-24.
42. Crone SG, Jacobsen A, Federspiel B, Bardram L, Krogh A, Lund AH, Friis-Hansen L. microRNA-146a inhibits G protein-coupled receptor-mediated activation of NF-κB by targeting CARD10 and COPS8 in gastric cancer. Mol Cancer 2012;11:71.
43. Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010;142:914-29.
44. Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, Sun G, Tay J, Linsley PS, Baltimore D. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med 2011;208:1189-201.
45. Rebane A, Runnel T, Aab A, Maslovskaja J, Rückert B, Zimmermann M, Plaas M, Kärner J, Treis A, Pihlap M, Haljasorg U, Hermann H, Nagy N, Kemeny L, Erm T, Kingo K, Li M, Boldin MP, Akdis CA. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J Allergy Clin Immunol 2014;134:836-47.e11.
46. Ying S, Taborda-Barata L, Meng Q, Humbert M, Kay AB. The kinetics of allergen-induced transcription of messenger RNA for monocyte chemotactic protein-3 and RANTES in the skin of human atopic subjects: relationship to eosinophil, T cell, and macrophage recruitment. J Exp Med 1995;181:2153-9.
47. Islam SA, Chang DS, Colvin RA, Byrne MH, McCully ML, Moser B, Lira SA, Charo IF, Luster AD. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat Immunol 2011;12:167-77.
48. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006.
49. Wang G, Tam LS, Li EK, Kwan BC, Chow KM, Luk CC, Li PK, Szeto CC. Serum and urinary free microRNA level in patients with systemic lupus erythematosus. Lupus 2011;20:493-500.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Bhardwaj N. MicroRNAs in atopic dermatitis: a review. J Transl Genet Genom 2017;1:15-22. http://dx.doi.org/10.20517/jtgg.2017.01
AMA Style
Bhardwaj N. MicroRNAs in atopic dermatitis: a review. Journal of Translational Genetics and Genomics. 2017; 1: 15-22. http://dx.doi.org/10.20517/jtgg.2017.01
Chicago/Turabian Style
Bhardwaj, Neeti. 2017. "MicroRNAs in atopic dermatitis: a review" Journal of Translational Genetics and Genomics. 1: 15-22. http://dx.doi.org/10.20517/jtgg.2017.01
ACS Style
Bhardwaj, N. MicroRNAs in atopic dermatitis: a review. J. Transl. Genet. Genom. 2017, 1, 15-22. http://dx.doi.org/10.20517/jtgg.2017.01
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 4 clicks
Cite This Article 4 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.