Riboflavin metabolism: role in mitochondrial function
Abstract
Riboflavin, known as vitamin B2, a water-soluble vitamin, is an essential nutrient in vertebrates, hence adequate dietary intake is imperative. Riboflavin plays a role in a variety of metabolic pathways, serving primarily as an integral component of its crucial biologically active forms, the flavocoenzymes flavin adenine dinucleotide and flavin mononucleotide. These flavocoenzymes ensure the functionality of numerous flavoproteins including dehydrogenases, oxidases, monooxygenases, and reductases, which play pivotal roles in mitochondrial electron transport chain, β-oxidation of fatty acids, redox homeostasis, citric acid cycle, branched-chain amino acid catabolism, chromatin remodeling, DNA repair, protein folding, and apoptosis. Unsurprisingly, impairment of flavin homeostasis in humans has been linked to various diseases including neuromuscular and neurological disorders, abnormal fetal development, and cardiovascular diseases. This review presents an overview of riboflavin metabolism, its role in mitochondrial function, primary and secondary flavocoenzyme defects associated with mitochondrial dysfunction, and the role of riboflavin supplementation in these conditions.
Keywords
Introduction
Riboflavin is the precursor of metabolically active flavocoenzymes which are utilized as cofactors for approximately 90 flavoproteins in numerous enzymatic reactions as either flavin adenine dinucleotide (FAD) (84%) or flavin mononucleotide (FMN) (16%)[1]. These flavoproteins (enzymes using flavocoenzymes) including dehydrogenases, oxidases, monooxygenases, and reductases play critical roles in mitochondrial electron transport chain, mitochondrial and peroxisomal β-oxidation of fatty acids, citric acid cycle, redox homeostasis, nitric oxide synthases, and branched-chain amino acid (BCAA) catabolism[2,3]. They are also involved in chromatin remodeling, DNA repair, protein folding, apoptosis[2] biosynthesis or regulation of other essential cofactors and hormones, including coenzyme A, coenzyme Q, heme, pyridoxal 5’-phosphate, steroids, and thyroxine; and metabolism of other B vitamins (folate, pyridoxine, and niacin) and P450 enzymes[1,4].
Riboflavin, as with other water-soluble B vitamins, must be obtained through diet as mammals have lost the ability to synthesize this molecule. Endogenous synthesis of riboflavin by microflora in the large intestine may contribute but to a lesser extent[5]. Hence, adequate dietary intake from major sources such as milk and dairy products, eggs, seafood, poultry, lean meat, cereals, and vegetables is imperative. The recommended daily allowance of riboflavin on average is 1.3 mg/day for adult men and 1.1 mg/day for women, with variations depending on age and reproductive status including pregnancy and lactation[5].
Riboflavin metabolism and transport
Riboflavin ingested in diet exists either as free riboflavin, predominantly found in milk and eggs, or its protein bound form as flavoproteins including FAD and FMN, which must be released from the carrier proteins to which they are bound. The latter occurs through dietary protein denaturation in the stomach and subsequent hydrolysis to free riboflavin by alkaline phosphatases and FMN/FAD pyrophosphatases in the ileal brush border to be absorbed in the small intestine[6][Figure 1]. Next, free riboflavin is transported into the enterocytes via carrier-mediated uptake by RFVT3 (previously hRFT2, encoded by SLC52A3), which functions primarily to absorb riboflavin from dietary intake[7]. This saturable uptake process occurs at the apical membrane and is reported to be linear up to approximately 30 mg riboflavin per meal[8], following which little additional absorption of riboflavin occurs[9,10].
Figure 1. Metabolism and transport of riboflavin and flavocoenzyme. Dietary FAD and FMN are converted to riboflavin by non-specific hydrolases on the brush-border membrane of ileal enterocytes and are subsequently absorbed into the intestine via apically expressed RFVT3. Inside the enterocytes, riboflavin can either be further metabolized to FMN by riboflavin kinase and subsequently to FAD by FAD synthase or released into portal blood by basolaterally expressed RFVT1 and RFVT2. Circulating plasma riboflavin associates with albumin or globulins or is converted into a coenzyme form in erythrocytes or leukocytes. RFVT2-mediated transport allows riboflavin uptake into the brain where it is highly expressed, and additionally into endocrine organs, such as pancreas, liver, and muscle tissue. The mechanism of import of riboflavin into the mitochondrial matrix has not been precisely elucidated to date. “RFVT?” is depicted as a putative riboflavin transporter responsible for this step. The mitochondrial FADT imports FAD from the cytosol into the mitochondria. The question mark indicates that FADT-mediated efflux of FAD from the mitochondrial matrix to the cytosol remains to be established (see also Barile et al.[5] 2016 and Balasubramaniam et al.[3] 2019). FAD: flavin adenine dinucleotide; FMN: flavin mononucleotide; FADT: flavin adenine dinucleotide transporter
After cellular uptake, free riboflavin undergoes adenosine triphosphate (ATP)-dependent phosphorylation by riboflavin kinase (RFK) (EC 2.7.1.26), a ubiquitous rate-limiting flavokinase, to form FMN, which is consequently adenylated to FAD by FAD synthase (FADS) (EC 2.7.7.2). Riboflavin may subsequently be released into the portal blood and to the liver in its free form or as FMN after being transported by RFVT1 (previously hRFT1) and RFVT2 (previously hRTF3), encoded by SLC52A1 and SLC52A2, respectively, and embedded within the basolateral membrane of the enterocytes[5]. Apart from being expressed in the gastrointestinal system, RFVT1 is also detected in the placenta where it transports maternal riboflavin to the fetus. RFVT2-mediated transport allows riboflavin uptake into the brain where it is highly expressed, and additionally in endocrine organs such as the pancreas, liver, and muscle[7].
Circulating plasma riboflavin is either bound to albumin and immunoglobulins or is converted into its coenzyme forms in erythrocytes or leukocytes. Unbound flavins are rapidly hydrolyzed to free riboflavin and excreted in urine. There is little or no storage of riboflavin in the body; hence, any intake in excess of tissue requirements or which surpasses renal reabsorption is eliminated in the urine as riboflavin or its catabolites 7-alpha-hydroxy riboflavin, 10-hydroxyethylflavin, and lumiflavin[5,11]. As a result, riboflavin has a relatively low toxicity even at supra-pharmacological doses[5].
Mitochondria-the cell’s powerhouse
Mitochondria are maternally inherited multifunctional double-membrane, highly dynamic cytoplasmic organelles, ubiquitously present in all cells, except erythrocytes. The mitochondrion is composed of several compartments that carry out specialized functions. These include the outer membrane, inner membrane, the intermembrane space, and cristae, which are the in-folding of the inner membrane and matrix. Mitochondria execute myriad vital cellular processes including fatty acid oxidation, urea cycle, Krebs cycle, biosynthesis of heme and steroids, maintenance of calcium homeostasis, caspase-dependent apoptosis, reactive oxidant species (ROS) generation, and heme and steroid synthesis[12,13]. In addition to these functions, the hallmark of mitochondria is the pivotal role it plays in aerobic cellular energy generation via oxidative phosphorylation (OXPHOS).
Riboflavin is a key component of the mitochondrial respiratory chain (RC), hence is pivotal for ATP generation and sustaining energy-dependent cellular functions. The OXPHOS system is a 5-enzyme complex which encompasses the mitochondrial respiratory chain (Complexes I-IV), Complex V, and two mobile electron shuttles (coenzyme Q10 and cytochrome c)[14][Figure 2]. Electrons derived from oxidation of pyruvate and fatty acids are transferred via NADH to Complex I (FMN-dependent NADH-ubiquinone oxidoreductase), while electrons from succinate in the Krebs cycle are transferred to Complex II (FAD-dependent succinate-ubiquinone oxidoreductase) via FADH2. Electrons are subsequently transferred to ubiquinone (Coenzyme Q10) and then to Complex III (reduced CoQ-cytochrome c reductase), and via cytochrome c to cytochrome c oxidase (COX) (Complex IV), the terminal oxidase of the RC before finally reducing molecular oxygen to water. The free energy liberated during this sequential electron transfer is used to generate an electrochemical gradient, by pumping protons from the matrix to the intermembrane space at three coupling sites (Complexes I, III, and IV). This proton gradient is used by Complex V (ATP synthase or F1F0 ATPase) to drive ATP synthesis from ADP and inorganic phosphate.
Figure 2. Schematic diagram of flavocoenzymes in mitochondrial energy metabolism. The OXPHOS system is a 5-enzyme complex which encompasses the mitochondrial respiratory chain (Complexes I-IV), Complex V, and two mobile electron shuttles (coenzyme Q10 and cytochrome c). Electrons derived from oxidation of pyruvate mediated by pyruvate dehydrogenase (PDH) and fatty acid oxidation are transferred via NADH to Complex I (FMN-dependent NADH-ubiquinone oxidoreductase), while electrons from succinate in the Krebs cycle, amino acid metabolism, and fatty acid oxidation are transferred to Complex II (FAD-dependent succinate-ubiquinone oxidoreductase) via FADH2. Electrons are subsequently transferred to ubiquinone (Coenzyme Q10) and then to Complex III (reduced CoQ-cytochrome c reductase), and via cytochrome c to cytochrome c oxidase (COX) (Complex IV), the terminal oxidase of the RC before finally reducing molecular oxygen to water. The free energy liberated during this sequential electron transfer is used to generate an electrochemical gradient of protons, which is finally used by Complex V (ATP synthase or F1F0 ATPase) to drive ATP synthesis from ADP and inorganic phosphate. RF: Riboflavin; RF kinase: riboflavin kinase; Q: Coenzyme Q10; CytC: cytochrome c oxidase; FMN: flavin mononucleotide; FAD: flavin adenine dinucleotide
Pathogenesis of riboflavin or flavocoenzyme deficiency and mitochondrial dysfunction
Riboflavin deficiency or defects in the production of its flavocoenzymes FAD and FMN can lead to disruption of the RC, consequently mitochondrial dysfunction, and increased production of ROS, overwhelming the cellular antioxidant mechanisms. Exposure to excessive ROS, including free radical superoxide O2-, hydrogen peroxide, and hydroxyl radicals, favors oxidative stress, alters mitochondrial Ca2+ homeostasis, triggers membrane lipid peroxidation and potentially induces nuclear and mtDNA damage[15].Sustained elevations in intracellular Ca2+ concentrations ultimately cause neuronal degeneration and cell death by initiating apoptosis[16]. All these perturbations are implicated in mitochondrial dysfunction, hence strategies to mitigate this may pose an important therapeutic avenue.
Classification of defects in riboflavin metabolism associated with mitochondrial dysfunction
Simplistically classified in this review, the primary defects of flavocoenzyme metabolism include disorders of riboflavin or flavocoenzyme transport and enzymes in the synthetic pathway of FMN and FAD. Secondary flavoproteome defects include FMN- and FAD-dependent reactions resulting in functional disruption of the cellular flavoproteins, but do not arise as a direct result of defects in the synthesis and transport of riboflavin, FMN, or FAD.
Secondary flavoproteome defects can be further subcategorized into primary mitochondrial disorders (PMD) and secondary mitochondrial dysfunction (SMD)[17]. PMD are genetic disorders that directly impair OXPHOS proteins or its function by impacting the complex machinery involved in the OXPHOS process. SMD can be caused by germline mutations in non-OXPHOS genes, accompany various hereditary non-mitochondrial diseases, or may be acquired secondary to adverse environmental factors which can cause oxidative stress. A summary of disorders of flavocoenzymes and flavoproteins associated with primary and secondary mitochondrial dysfunction is presented in Table 1.
Overview of disorders of flavocoenzymes and flavoproteins associated with primary and secondary mitochondrial dysfunction
| Disease name | Gene | Phenotype OMIM number | Primary clinical phenotype | Biochemical findings | Riboflavin responsiveness |
|---|---|---|---|---|---|
| Riboflavin transporter deficiency | SLC52A2 | 614707 | Sensorimotor and cranial
neuropathy Sensorineural hearing loss | Multiple acyl Co-A dehydrogenase defect (MADD) profile on acylcarnitine and ethylmalonic aciduria | Yes |
| SLC52A3 | 211500 | Sensorimotor and cranial
neuropathy | |||
| 211530 | Sensorimotor and cranial neuropathy
Sensorineural hearing loss | ||||
| FAD transporter deficiency | SLC25A32 | 616839 | Neuromuscular e.g. exercise intolerance, ataxia, muscle weakness | MADD profile
Complex II deficiency | Yes |
| FAD synthase deficiency | FLAD1 | 255100 | Fatal infantile onset hypotonia, swallowing difficulties, respiratory insufficiency, cardiomyopathy
Late onset lipid storage myopathy | MADD profile
Combined respiratory chain deficiencies | Yes |
| Mitochondrial complex I deficiency, nuclear type 20 | ACAD9 | 611126 | Early onset and lethal hypertrophic cardiomyopathy, encephalopathy | Complex I deficiency | Yes |
| Later presentation with myopathy, e.g., exercise intolerance and muscle weakness | |||||
| Mitochondrial complex I deficiency, nuclear type 19 | FOXRED1 | 618241 | Leigh syndrome
Encephalomyopathy | Complex I deficiency | Limited data (n = 2) |
| COQ10 deficiency-6 | COQ6 | 614650 | Steroid resistant nephrotic syndrome, sensorineural hearing loss | COQ10 deficiency | No data |
| Myopathy, mitochondrial progressive, with congenital cataract, hearing loss, and developmental delay | GFER | 613076 | Neuromuscular e.g. hypotonia, muscle weakness, psychomotor retardation, muscle weakness, Cataracts, hearing loss | Isolated Complex IV deficiency or combined deficiencies | No data |
| Auditory neuropathy and optic atrophy | FDXR | 617717 | Auditory neuropathy and optic atrophy, encephalopathy | Combined respiratory chain deficiencies | No data |
| Mitochondrial Complex I deficiency, nuclear type 4 | NDUFV1 | 618225 | Encephalopathy, motor delay
Neurodevelopmental regression, Oculomotor impairment | Complex I deficiency | Limited data (n = 7), riboflavin administered with other supplements[94,97,98,101] |
| Mitochondrial Complex I deficiency, nuclear type 7 | NDUFV2 | 618229 | Hypertrophic cardiomyopathy
Encephalopathy Leigh syndrome | Complex I deficiency | Limited data (n = 1), riboflavin administered with other supplements[113] |
| Mitochondrial Complex II deficiency | SDHA | 252011 | Leukoencephalopathy, Leigh syndrome, cardiomyopathy, Cancer susceptibility (gastrointestinal stromal tumours, paraganglioma/pheochromocytoma, pituitary adenoma and renal carcinoma) | Complex II deficiency | No response (n = 2)[133] |
| Multiple acyl-coenzyme A dehydrogenase deficiency (MADD) | ETFA, ETFB, ETFDH | 231680 | Neonatal onset- with (type I) or without congenital anomalies (type II) and
early onset encephalopathy and cardiomyopathy Late onset myopathy (type III) | Increase in short, medium and long chain acylcarnitines
Characteristic MADD pattern in urinary organic acids | YES - established for late onset myopathy[138] |
| Dihydrolipoamide dehydrogenase (E3) deficiency | DLD | 246900 | Early onset encephalopathy, Primary liver involvement, Myopathy
| Elevated lactate, pyruvate, alpha ketoglutarate, branched chain amino acids and alpha ketoacids | YES - established for myopathic form[146] |
| Ethylmalonic encephalopathy | ETHE1 | 602473 | Encephalopathy, Vasculopathy (petechial purpura, orthostatic acrocyanosis, chronic hemorrhagic diarrhoea) | Increase in ethylmalonic, C4 and C5 acylcarnitines.
Complex IV deficiency | Limited data (n = 4)[152] |
| Combined oxidative phosphorylation deficiency 6 | AIFMI | 300816 | Encephalomyopathy | Combined respiratory chain deficiencies | Limited data (n = 2)
for ataxia[160] and neurological condition (n = 1, gross, fine motor and communication)[155] |
| Cowchock syndrome | AIFMI | 310490 | X-linked Charcot-Marie-Tooth disease (CMTX4) with axonal sensorimotor neuropathy, deafness and cognitive Impairment | ||
| Deafness, X-linked 5 | AIFMI | 300614 | Auditory neuropathy with peripheral neuropathy | ||
| Spondyloepimetaphyseal dysplasia, X-linked, with hypomyelinating leukodystrophy | AIFMI | 300232 | Spondyloepimetaphyseal dysplasia with hypomyelination |
Primary disorders of flavocoenzyme metabolism associated with mitochondrial dysfunction
Disorder of Riboflavin Transport
Riboflavin Transporter Deficiency Neuronopathy (OMIM #614707, OMIM# 211500, and OMIM #211530) – previously known as Brown-Vialetto-Van Laere and Fazio- Londe syndrome
Human riboflavin transporters, RFVT1, RFVT2, and RFVT3, are encoded by their respective genes, SLC52A1, SLC52A2, and SLC52A3[18-21]. These transporters with different tissue expressions transport riboflavin across plasma membranes and maintain the supply of flavins to the cells.
Mutations in the SLC52A2 and SLC52A3 have been reported to cause Brown-Vialetto-Van Laere (BVVL) and Fazio-Londe (FL)[22,23]. Both conditions are now considered as a single disease entity[24], with progressive sensorimotor and cranial neuropathy in both and absent sensorineural hearing loss in FL as primary manifestations. A new nomenclature was proposed to clarify the specific disease mechanism and both conditions were renamed to riboflavin transporter deficiency (RTD)[25], with mutations in SLC52A2 and SLC52A3 causing RTD2 and RTD3, respectively[26]. A recent review identified 109 RTD patients (52 RTD2, 56 RTD3, and 1 RTD2/3)[26]. The overall mean age of onset of these patients was 5.3 years (range: 0-5 years). The predominant clinical features were hearing loss and muscle weakness. Some characteristics of patients with RTD3 included a later onset of presentation (> 10 years to third decade) and bulbar symptoms. The common presenting features of patients with RTD2, on the other hand, was abnormal gait and/or ataxia[26]. Plasma acylcarnitine profiles prior to riboflavin supplementation was suggestive of multiple acyl-CoA dehydrogenase defect (MADD)[22]. Urine organic acids commonly showed ethylmalonic aciduria suggesting impaired fatty acid, methionine, and/or isoleucine oxidation[26]. Both plasma acylcarnitine and urine organic acids, however, have been observed to be normal in nearly half of RTD patients[26].
Effective doses of riboflavin varied 10-80 mg/kg/day with over 70% of patients demonstrating improvements in muscle strength, motor abilities, respiratory function, and/or cranial nerve deficits[26]. The pathomechanism of the specific vulnerability of neurons in RTD might be due to mitochondrial dysfunction and impairment in the clearance of reactive oxygen species[26].
Haploinsufficiency of the SLC52A1 (OMIM#615026) due to maternal microdeletion and heterozygous intronic variant has been reported to cause a transient riboflavin responsive neonatal multiple acyl-CoA dehydrogenase deficiency that resolved with oral supplementation of riboflavin[27-29].
Disorders of Flavocoenzyme Transport
Mitochondrial FAD transporter deficiency (OMIM #616839)
SLC25A32 encoding the mitochondrial FAD transporter has been described in two patients with biallelic mutations to date[30,31]. The first, a 14-year-old girl presented with riboflavin-responsive recurrent exercise intolerance and biochemical findings of MADD[30]. The second, a 51-year-old Dutch patient with a severe neuromuscular phenotype had initially presented aged three years with muscle weakness post- influenza infection. He subsequently developed impaired motor skills, progressive exercise intolerance in childhood, early-onset ataxia, myoclonus, dysarthria, and dysphagia[31]. Muscle biopsy in both patients demonstrated ragged-red fibers, lipid storage, and decreased staining for succinate dehydrogenase (SDH, FAD-dependent mitochondrial respiratory chain Complex II) and COX (mitochondrial respiratory chain Complex IV). Complex II deficiency was revealed in cultured skin fibroblasts and muscle from the first and second patients, respectively[30,31]. Dramatic improvements in exercise tolerance and endurance and biochemical abnormalities were reported in both patients following oral riboflavin supplementation[30,31].
SLC25A32 has recently been revealed to be a novel regulator of cancer cell proliferation and mitochondrial FAD metabolism. SLC25A32 knock-down in sensitive tumor cells resulted in inhibition of the FAD-dependent Complex II, increased succinate levels, and reduced oxygen consumption rate[32]. These findings corroborate evidence of decreased Complex II protein levels and OXPHOS activity, which is a marker for mitochondrial FAD in muscle of patients with severe neuromuscular phenotype and novel variants in SLC25A32[30,31]. Reduction of mitochondrial FAD concentrations by inhibition of SLC25A32 is anti-proliferative in a subset of tumor cell lines and has potential clinical applications as a novel cancer target by increasing oxidative stress and reducing tumor growth[32].
Disorders of Flavocoenzyme Metabolism
Flavin adenine dinucleotide synthase (FAD) synthase deficiency (OMIM # 255100)
The FLAD1 gene encodes FAD synthase (EC 2.7.7.2), which catalyzes the adenylation of FMN into the redox cofactor FAD. Human FADS has previously been shown to be a bifunctional enzyme with both FAD synthase and hydrolase activity[33]. The enzyme contains an N-terminal molybdopterin binding (MPTb) domain, which has FAD hydrolase activity, and a C-terminal 3’-phosphoadenosine-5’-phosphosulfate (PAPS) reductase domain, which is sufficient to catalyze FAD synthesis and has thus been renamed the FADS domain[33,34]. Multiple FADS isoforms generated due to alternative splicing of the encoding FLAD1 gene result in distribution within different subcellular localizations[34-36].Two isoforms, the more abundant cytosolic (FADS2) and the mitochondrial (FADS1), have been described in detail[33]. Human FADS1, FADS2, and FADS3 possess both catalytic domains and are bifunctional, while FADS4 and FADS5 present the sole mMPTb domain and are monofunctional enzymes[37]. A novel cytosolic FADS6 isoform recently characterized contains only the PAPS domain with considerable FAD synthesis activity, which allows survival of affected patients with biallelic FLAD1 frameshift variants[37].The FADS6 isoform has approximately 70% of the FAD-synthesizing capacity of human FADS2. Its catalytic efficiency (kcat/Km) is however higher due to a lower Km for FMN and ATP compared to FADS2[38].
The recently described clinical spectrum for FAD synthase deficiency has ranged from neonatal-onset, a lethal disease with metabolic myopathy, cardiomyopathy, swallowing, speech difficulties, and respiratory insufficiency, to later-onset, a potentially treatable neuromuscular disorder with lipid storage myopathy, MADD-like metabolic aberrations, and combined respiratory-chain deficiency[34]. To date, sixteen patients have been reported with age of onset ranging from birth to 44 years[34,39-45]. Most patients (92%) have however presented in infancy, with five out of thirteen dying within the first twelve months of life. The first patient described by Taylor et al.[39], a Turkish male infant of consanguineous parents, presented at age four months with respiratory insufficiency, lipid myopathy, decreased activities of mitochondrial respiratory chain Complexes I and IV, and early death at eight months of age. A homozygous 4-bp deletion (c.397_400delTTCT) in the FLAD1 gene was identified, resulting in a frameshift and premature termination (Phe134CysfsTer8). Functional studies were not performed; however, in silico predictions of the deleterious effect of the mutations supported a causal association.
A further 15 patients from 13 unrelated families affected by MADD and/or multiple respiratory-chain deficiency and lipid storage myopathy were diagnosed with FLAD1 deficiency. Apart from one patient, all were identified by whole-exome sequencing or through candidate gene panels[3]. Characteristic clinical manifestations included early infantile onset hypotonia, swallowing, speech difficulties, respiratory insufficiencies, and a fatal course (seven died within the first year of life and one at age 16 years). Cardiorespiratory collapse was the cause of death for a neonate who presented at 32 h of age and died at three days. One patient died of multiorgan failure at seven months due to recurrent episodes of sudden cardiac arrest necessitating a pacemaker. Another infant with cardiomyopathy and recurrent supraventricular tachycardias who required implantable cardioverter defibrillator placement showed a dramatic response to riboflavin supplementation and was still alive at 22 years. Interestingly, the acute metabolic decompensations observed in the neonatal forms of MADD caused by ETFA, ETFB, and ETFDH defects, including hypoglycemia, metabolic acidosis, and hyperammonemia have not been reported in early-onset FADS deficiency[41]. The adult onset presentations at 20 and 44 years included exercise intolerance and progressive muscle weakness, with gait difficulties, bilateral foot drop, and arm weakness in the older patient. These individuals were alive at 44 and 56 years, respectively[34,40].
Metabolic findings in FADS deficiency are typical of riboflavin deficiency or MADD, with increased acylcarnitines (C4-C14 even chain acylcarnitines; C5 and C5-DC; and C10:1, C14:1, C16:1, C18:1, and C18:2) and urinary organic acids (increased urinary excretion of ethylmalonic, adipic, suberic, glutaric and methylsuccinic acids, hexanoylglycine, and tiglylglycine)[3]. Characteristic muscle pathology with pronounced lipid storage and global decrease of COX and/or SDH histochemical staining has been reported[39]. Multiple respiratory chain enzyme deficiencies involving Complexes I-III were described in skeletal muscle biopsies in seven of eight cases tested[34,39,41].
A possible genotype-phenotype correlation has been proposed with homozygous carriers of frameshift variants affecting the MPTb domain displaying a more severe clinical course than that observed in individuals harboring single amino acid changes in the FADS domain that are potentially more responsive to riboflavin therapy[34].
Riboflavin supplementation resulted in clinical improvements in nine of ten patients treated[34,39-42,44,45]. All five children who presented in infancy and treated with riboflavin were alive, two at 8 years, one at 22 years, one at 15 months[44]), and one at 2 years 5 months[45]. Treatment with riboflavin in two patients carrying homozygous c.401_404delTTCT mutations in FLAD1 at age three months resulted in milder improvements in spontaneous activity, muscle tone, vomiting, and alertness; however, it failed to prevent disease progression and demise by six and five months, respectively[41]. The same variant had previously been reported in an untreated Turkish infant with multiple respiratory chain complex deficiencies who died at six months of age[34]. It has been proposed that the partial response to riboflavin supplementation and residual FADS activity described in patients with biallelic frameshift mutations in exon 2 of FLAD1 may be attributed to the existence of a FADS isoform that lacks exon 2, but has an intact and functional FADS domain[34]. However, the doses of riboflavin used in these patients may have been inadequate in ensuring long-term survival[41].
A trial of riboflavin therapy should be offered to all FADS-deficient individuals while awaiting mutational analysis. The benefits of early diagnosis and treatment is exemplified by the fatal outcome of the untreated brother of an older sister who responded to riboflavin[34].
Riboflavin kinase deficiency (OMIM # 613010)
RFK (EC 2.7.1.26) is a ubiquitous rate-limiting enzyme that catalyzes the first step in flavocoenzyme biosynthesis by phosphorylating riboflavin to form FMN, an obligatory step in riboflavin metabolism. FMN is subsequently adenylated by FAD synthetase to generate FAD[46]. Embryonic lethality demonstrated before Day 7.5 of gestation in complete knockout mice was potentially attributable to secondary mitochondrial dysfunction from the effect on flavocoenzyme deficiencies on the electron transport chain, which is critically involved in energy generation[47]. To date, there have been no reported human phenotypes of RFK deficiency. It is possible that defects in RFK may be incompatible with life due to its critical role noted above. Hypomorphic RFK mutations may result in clinical phenotypes that are not that dissimilar to patients with FLAD1 mutations[3].
Secondary flavoproteome defects associated with mitochondrial dysfunction
Primary Mitochondrial Disorders
Acyl-CoA dehydrogenase-9 deficiency (ACAD9) (OMIM #611126)
Acyl-CoA dehydrogenase 9 (ACAD9), a mitochondrial protein, was initially demonstrated to catalyze the initial rate-limiting step in the beta-oxidation of long-chain fatty acids[48]. It was subsequently proposed to be a novel assembly factor crucial for oxidative phosphorylation Complex I biogenesis, independent of its role in fatty acid oxidation[49]. More recently, human ACAD9 was showed to be a bifunctional enzyme involving primarily oxidative phosphorylation Complex I biogenesis, with an additional moonlighting function in fatty acid oxidation (FAO)[50].
Major clinical presentations of ACAD9 deficiency include biventricular hypertrophic cardiomyopathy (85%), muscle weakness (75%), exercise intolerance (72%) and lactic acidosis[51]. Two subgroups of ACAD9 deficient patients have been described, including early-onset, often lethal cardiac involvement presenting in infancy. Those surviving from this group performed more poorly than later-onset presentations. Interestingly, developmental delay and severe intellectual disability were only seen with early disease onset[51]. ACAD9 is the only long-chain ACAD enzyme expressed in the brain and is especially abundant during fetal life[49,52]. Hence, it has been proposed that the neurological symptoms present in the most severely affected ACAD9-deficient patients could be attributed to a deficiency in brain FAO. To date, no patients with two ACAD9 null mutations have been described, suggesting that the complete absence of the protein is lethal. Embryonic lethality was also proposed for the failure to generate homozygous knockout ACAD9 mice[49].
Riboflavin supplementation resulted in alleviation of symptoms in 65% patients, and, most notably, improved survival was observed when commenced within the first year of life[51]. Riboflavin responsiveness may be related to its essential function as a precursor of FAD cofactor for ACAD enzyme activity and stability. It also increases ACAD9 protein levels and rescues Complex I assembly, while also functioning as a chemical chaperone by improving folding of ACAD9 mutant proteins[49]. Schiff et al.[49] (2015) suggested that similar specific interventions used for VLCAD defects including avoidance of fasting, medium-chain triglycerides, or triheptanoin anaplerotic therapy could be beneficial in terms of long-term outcomes in patients.
FAD-dependent OXidoREDuctase deficiency (FOXRED1) (OMIM #618241)
FOXRED1 has been proposed to be a dual function protein. It plays a key role as an assembly factor for Complex I biogenesis, and, secondly, due to its oxidoreductase activity, it is hypothesized to participate in glycine metabolism which modulates glutathione biosynthesis, an antioxidant protecting the cells from ROS[53-55]. Co-immunoprecipitation experiments suggest that FOXRED1 facilitates Complex I assembly by associating with the 370-kDa subcomplex and two other FAD-dependent Complex I assembly factors, ACAD9 and probably AIFM1[55].
FOXRED1 mutations have been recognized as a cause of Complex I deficiency. To date, eight patients from six families have been described with variable clinical spectrum and severity[55-60]. Age of onset of clinical manifestations ranged from birth to early infancy with one prenatal onset of oligohydramnios, severe intrauterine growth retardation, and periventricular cysts[60]. The clinical manifestations included Leigh syndrome and infantile-onset encephalomyopathy with epilepsy, mild to severe psychomotor retardation, and hypotonia. Non-neurological features included congenital lactic acidosis, cardiomyopathy, hepatomegaly, kyphoscoliosis, optic atrophy, roving eye movements, strabismus, and distal renal tubular acidosis[55-58,60]. Brain neuroimaging (MRI) findings ranged from normal to delayed myelination and features consistent with Leigh syndrome[55-57]. Enzymatic studies supported Complex I deficiency in all patients. Additionally, a milder decrease in Complex II was observed in one patient[59]. The majority of mutations identified were missense. Riboflavin supplementation was reported in only two patients[57,60], of whom one had prenatal onset and died at three months of age[60]. Outcome was available in 6/8 patients: 4/6 are alive with median age of 17 years (age range: 10-22 years) and another died at eight years. In contrast to patients with other nuclear encoded Complex I defects who typically have a severe clinical presentation and associated early death, it has been suggested that pathogenic variants in the FOXRED1 gene result in partial loss of function and are probably hypomorphic due to the longer survival of patients[61].
COQ6 deficiency (OMIM #614650)
Human Coenzyme Q10 monooxygenase 6 (COQ6) is a flavoprotein involved in the biosynthesis of Coenzyme Q10 which operates as a redox carrier by transferring electrons from respiratory chain Complexes I and II to Complex III. Coenzyme Q10 is also a potent antioxidant[62] and a cofactor of many mitochondrial dehydrogenases. It is required for pyrimidine nucleoside biosynthesis and has been implicated in the inhibition of apoptosis by preventing the collapse of the inner mitochondrial membrane[63]. Studies using human cell line lacking functional COQ6 showed impaired COQ synthesis, severe ATP deficiency, and increased production of reactive oxygen species[64].
In 2011, Heeringa et al.[65] reported a novel cause for steroid-resistant nephrotic syndrome associated with sensorineural hearing loss due to variants in the COQ6 gene. Twenty-three patients in 16 families with similar clinical manifestations and two mutations in the COQ6 gene have been identified to date[65-71]. Extra-renal manifestations such as ataxia, seizures, muscle weakness, white matter changes, and optic atrophy were less common[65,68]. The age of onset of the renal disease varied from 2 months to 6.4 years but was most common in infancy and early childhood. The course was progressive with median age of onset of end stage renal disease between 13 months and 19 months[65,68]. The most common histopathologic finding was focal segmental glomerulosclerosis. Abnormal mitochondrial proliferation in the podocytes was also noted on electron microscopy[68].
Coenzyme Q10 supplementation alone at 5-30 mg/kg/day was reported in a few patients. Significant improvement in proteinuria with normal renal function was noted on follow-up of these patients[66,70,72-74]) However, hearing loss had not improved after Coenzyme Q10 supplementation[66,72]. Hence, early supplementation of Coenzyme Q10 seemed to help avoid development of massive proteinuria and consequently chronic renal failure[72].
GFER deficiency (OMIM #613076)
Human Growth Factor ERV-1-like (GFER) (also known as augmenter of liver regeneration 1, ALR1) is one of the sulfhydryl oxidases with a FAD-binding domain, which together with disulfide carrier Mia40 forms part of the disulfide relay system (DRS) and constitutes the mitochondrial import and assembly (MIA) pathway. The MIA pathway plays an important role in the biogenesis of many mitochondrial intramembrane space (IMS) proteins[75]. GFER has two distinct isoforms: the long isoform (205 amino acids, 23 kD), which is mainly located in the IMS of the mitochondria, and the shorter isoform (125 amino acids, 15 kD), which is present in the nucleus. Mia40 functions as an import receptor of cysteine-rich substrates in the IMS and promotes oxidative folding of these proteins. It is re-oxidized by GFER in a disulfide-transfer reaction with electrons transferred to cytochrome c, thus connecting the DRS to the electron-transport chain of the mitochondria and establishing its role in Complex IV activity[76].
In 2009, Di Fonzo et al.[77] reported homozygous missense mutations (c.581 G > A p.R194H) in the GFER gene in three siblings from a consanguineous Moroccan family. Subsequent functional studies using analogous mutation (R182H) in yeast ERV1 showed altered mitochondrial morphology, mitochondrial instability, reduced cyt C oxidase activity, weaker FAD binding, decreased thermal protein stability, and altered protein folding[77,78]. Daithanka et al.[79] (2010) also characterized the human GFER/ALR protein and demonstrated that the R194H led to marked loss of protein stability by an increased rate of FAD dissociation and enhanced proteolysis susceptibility with only minimal effect on enzymatic activity.
There are eight patients in four families reported with mutations in the GFER gene[77,80,81], to date. The age range of the patients was 3 to 21 years with one deceased at 21 years of age. The predominant features were neuromuscular including hypotonia and psychomotor retardation of variable severity, muscle hypotrophy, progressive muscle weakness requiring ventilatory support, and, additionally, cataracts and lactic acidosis. Other clinical features included movement disorder, dysautonomia, cachexia and orthopedic problems[81], hearing loss[77], and adrenal insufficiency[80]. MRI of the brain showed normal findings (4/6), cerebellar and moderate cortical atrophy (1/6), and thin corpus callosum (1/6). Histopathologic muscle findings included abnormal mitochondrial morphology, ragged red fibers, COX negative, and atrophic fibers. Enzymatic testing revealed isolated Complex IV deficiency or combined deficiencies of Complexes I-IV. The missense mutation (c.581 G > A; p.R194H) was identified in six out of the eight patients.
Riboflavin supplementation was not reported in these patients. Experiments have indicated that the FAD cofactor is detached from Erv1 R182H during the catalytic reaction and inactivated the protein[78]. The functional defect was shown to recover by adding extra FAD[78]. Hence, riboflavin supplementation in patients with GFER mutations may potentially have clinical benefit, but this is speculative.
FDXR deficiency (OMIM #617717)
FDXR (Ferredoxin Reductase) has one FAD/NAD(P)-binding domain and two NAD(P)-binding domains. It is the sole ferredoxin reductase in humans and has important roles in iron-sulfur (Fe-S) cluster biogenesis and heme synthesis[82]. Mitochondrial membrane associated FDXR reduces the ferredoxins, FDX1 and FDX2, and transfers electrons from NADPH to the mitochondrial cytochrome P450 system, thus initiating the mitochondrial electron transport chain reaction[83]. Studies using patient derived fibroblasts showed loss of FDXR function led to decreased enzyme activity, reduced functions of Complexes I-III, significant increase in reactive oxygen species production, and mitochondrial iron overload[84,85].
There are 27 patients in 19 families with mutations in the FDXR gene reported in the literature[84-86]. A cohort of eight patients presented predominantly with auditory neuropathy and optic atrophy with onset in childhood or adolescence[84]. There were no other clinical features except for mild language delay in one and retinitis pigmentosa, ophthalmoplegia, and lower limbs hypoallesthesia in another patient[84]. However, clinical variability was present in another cohort of patients who presented with neurological features of early onset, i.e., infancy to childhood[85]. Apart from visual problems, i.e., retinitis pigmentosa and/or optic atrophy, and hearing loss, this cohort of 13 unrelated patients also had other clinical features such as hypotonia, global developmental delay, axonal sensorimotor polyneuropathy, regression after febrile illness, ataxia, failure to thrive, progressive microcephaly, and encephalopathy[85]. Respiratory chain enzyme activities, when reported, showed normal activities or combined deficiencies. Post-mortem findings of one FDXR patient showed neuronal loss and vacuoles in the cerebral cortex and increased biomarkers for gliosis, astrocyte activation, and neurodegeneration[86]. These findings were also observed in a homozygous p. R389Q mouse model, suggesting that inflammation could be a major component of the pathology in the neurodegeneration of FDXR mutations[86]
NDUFV1 deficiency (OMIM # 618225)
NADH: ubiquinone oxireductase flavoprotein 1 (NDUFV1) is a 51-kDa highly conserved nuclear encoded subunit comprising the electron input (N) functional module of Complex I[87]). It contains NADH-, FMN-, and Fe-S-binding sites[88].
Mutations in the NDUFV1 gene causing Complex I deficiency have been identified in 41 patients in 33 families[56,88-110]. The median age of onset of clinical manifestations was 8.0 months (range: birth to 6 years). The most common clinical features were motor delay or neurodevelopmental regression usually precipitated by an illness, hypotonia, dystonia, seizures, spasticity, and cognitive impairment. Oculomotor impairment including ptosis, strabismus, and ophthalmoplegia was also frequently observed. Typical Leigh syndrome[88,107]and late-onset Leigh syndrome have been reported[95,103]. The clinical course was variable with some patients having slower progression of neurological manifestations[90,91,103]. Brain MRI and MRS findings included features of typical Leigh syndrome, white matter changes, basal ganglia changes, elevated lactate peak, infantile striatal necrosis[97], and cavitating leukoencephalopathy[109]. Outcome was available in 34 patients; ten died at a median age of six months (range: three days to three years). The median age among surviving patients was seven years (range: 1-15 years) and severity of neurocognitive outcome varied from mild learning disability[91,96] to significant impairment[101,103]. Riboflavin supplementation was only reported in seven patients[94,97,98,101] and was given with other supplements such as thiamine, Coenzyme Q10, creatine, and alpha lipoic.
NDUFV2 deficiency (OMIM #618229)
Human NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2), a 24-kDa binuclear [2Fe-2S] cluster containing protein, is highly conserved and one of the nuclear encoded subunits of Complex I which is involved in electron transfer. It is also one of the subunits comprising the electron input (N) functional module of Complex I[87].
Phenotypic variability is observed in patients with Complex I deficiency due to mutations in the NDUFV2 gene. Bénit et al.[111] 2003 reported three affected siblings in a consanguineous family presenting with early onset hypertrophic cardiomyopathy, truncal hypotonia, feeding difficulties, growth retardation, and early death within the first year of life. Another patient also presented with hypertrophic cardiomyopathy and encephalopathy[112]. However, a separate family with three affected siblings presented with Leigh syndrome without cardiac involvement[113]. These patients reported by Cameron et al.[113] had seizures by 10 months of age, progressive clinical course, and two died at 19 months and 10 years. One of these patients was started on riboflavin plus other supplements. One sibling is still alive at 32 years despite a vegetative state by three years of age. All these patients were homozygous for a 4-bp deletion in intron 2 (IVS2 + 5_ + 8delGTAA) of the NDUFV2 gene[113]. Studies using human disease cell model showed that this deletion caused a significant reduction in the mitochondrial targeting ability of the NDUFV2 protein[114].
In a cohort of 37 children with cavitating leukoencephalopathies, three (3/37, 8.1%) were compound heterozygotes for pathogenic mutations in the NDUFV2 gene[109]. The age of onset of symptoms was 4-46 months with stable/improved clinical course and frontal predominant and deep white matter patterns on brain MRI[109].
Succinic dehydrogenase subunit A deficiency (OMIM #252011)
Succinic dehydrogenase subunit A (SDHA), a flavoprotein with FAD as cofactor, is a nuclear encoded and catalytic subunit of Complex II (succinate dehydrogenase) which oxidizes succinate to fumarate in the Kreb’s cycle and transfers electrons to ubiquinone in the mitochondrial electron transport chain[115].
Mutations in SDHA gene result in both a mitochondrial disease causing Complex II deficiency and tumor susceptibility. A large proportion (47%) of germline mutations in SDHA has been reported in gastrointestinal stromal tumors[116]. Other tumors such as paraganglioma/pheochromocytoma, pituitary adenoma, and renal carcinoma have also demonstrated SDHA germline mutations[117-121].
Complex II deficiency due to mutations in SDHA presents with variable clinical presentations and age of onset from prenatal to adulthood. The first report of a nuclear gene mutation causing a mitochondrial respiratory chain deficiency in humans was identified in two sisters presenting with developmental regression at 10 months of age, pyramidal tract signs, leukodystrophy with early demise, Complex II deficiency, and homozygous mutations (R554W) in the SDHA gene[122,123]. Further reports of patients with SDHA mutations have since been described[124-131]. Leigh or Leigh-like syndrome was a common phenotype. Slower progression with survival up to 11 years of age has been observed[126,127,130], and, in one patient, there was no apparent cognitive impairment at 10 years of age[130]. Isolated cardiomyopathy presenting from 32 weeks in utero to 10 years of age was associated with high mortality in two large Bedouin families with homozygous G555E mutations in the SDHA gene[132].
Adult presentation has been reported in two sisters (62 and 56 years) with normal cognition, late-onset (mid-40s), and slowly progressive cerebellar ataxia, visual impairment, optic atrophy, proximal weakness, and partial Complex II deficiency in skeletal muscle[133]. Riboflavin supplementation did not prevent neurological progression in these two patients. These patients were found to be heterozygous for a missense mutation (R408C) in a highly conserved region of the mature SDHA protein, suggesting an autosomal dominant inheritance pattern[134]. Another family with three affected members who presented with optic atrophy, dilated cardiomyopathy, ataxia, and isolated Complex II deficiency in fibroblast were also found to be heterozygous for a R451C missense mutation. This family serves as the second report supporting an autosomal dominant inheritance[135].
Secondary mitochondrial dysfunction
Multiple Acyl-CoA Dehydrogenase Deficiency (OMIM# 231680)
Multiple acyl-coenzyme A dehydrogenase deficiency, also known as glutaric aciduria Type II (GAII), is an autosomal recessive disorder that affects the oxidation of fatty acids, BCAA, lysine, tryptophan, and choline. MADD is caused by deficiency of one of the two electron-transfer flavoproteins which transfer electrons from acyl-CoA dehydrogenases to Coenzyme Q in the respiratory chain: Electron Transfer Flavoprotein (ETF), encoded by ETFA and ETFB genes, and electron-transfer flavoprotein dehydrogenase (ETFDH), encoded by ETFDH[136]. Metabolic defects resulting from impaired beta-oxidation include decreased ATP biosynthesis, excessive lipid accumulation in various organs, and insufficient gluconeogenesis[137].
The clinical phenotype is variable and has been classified into neonatal onset forms, with the most severely affected patients presenting with congenital anomalies (Type I) or without anomalies (Type II), and mild and/or later onset (Type III)[138]. Severely affected patients present in the first few days of life with non-ketotic hypoglycemia, hyperammonemia, and metabolic acidosis accompanied by hypotonia, encephalopathy, hepatomegaly, cardiomyopathy, and poor prognosis. An odor of sweaty feet similar to that in isovaleric acidemia may be observed[139]. Some patients have congenital anomalies (including large cystic kidneys, hypospadias, and neuronal migration defects that can be prenatally detected by fetal MRI and facial dysmorphism (low set ears, high forehead, and midfacial hypoplasia)[139]. The most frequent clinical presentation is the milder myopathic Type III form, which manifests with fluctuating proximal and axial myopathy with exercise intolerance and occasionally respiratory insufficiency[138] or rhabdomyolysis, although often with hepatomegaly, encephalopathy, and episodic lethargy, as well as vomiting and hypoglycemia often triggered by metabolic stress, episodes of which have been lethal in 5% of patients[140]. Although most decompensations occur in childhood, severe metabolic crises have also been reported in adulthood[138]. The majority of the 350 cases of late-onset MADD described in the literature carry mutations in the ETFDH gene (93%), while mutations in the ETFA (5%) and ETFB (2%) genes are less frequent[138].
Laboratory parameters include increased creatine kinase and lactate levels, with low carnitine. Diagnostic confirmation is made by the increase of short-, medium-, and long-chain acyl-carnitines on acylcarnitine analysis and characteristic urinary organic acid pattern comprising elevated levels of glutaric, ethylmalonic, 3-hydroxyisovaleric, 2-hydroxyglutaric, 5-hydroxyhexanoic, and relevant ketonuria, particularly glycine conjugates of C4 and C5 acids[137,138]. Muscle biopsies usually reveal lipid storage myopathy and secondary mitochondrial dysfunction with decreased Complexes I and II + III, attributable to deficiency of Coenzyme Q, which has been associated with increased ROS generation due to electron leak from misfolded variant ETFDH proteins and impaired Q10 binding affinity[138].
A clear genotype-phenotype correlation has been reported with the heterogenous subtypes of MADD. Type I disease is usually associated with homozygosity for null mutations. Even minor amounts of residual ETF/ETFDH activity suffices to prevent embryonic development of congenital anomalies observed in Type II disease, whereas higher residual activity is found in the late-onset form/ Type III disease. The genotype-phenotype correlation within Type III patients is however poor, as exogenous stressors including febrile infections may modulate the residual activity[138].
Riboflavin supplementation has successfully ameliorated clinical symptoms and metabolic abnormalities in almost all patients (98%) with late-onset MADD, the majority of whom have ETFDH mutations (93%)[138]. High levels of FAD and FMN due to riboflavin supplementation promote folding and stability of flavoproteins, particularly at fever simulating temperatures (40 °C), permitting certain mutant flavoenzymes to reach a folding efficiency and stability that attenuates the enzyme deficiency[7]. Combined treatment of riboflavin and Coenzyme Q10 has been advocated for use in riboflavin-responsive MADD based on studies which demonstrated improved mitochondrial dynamics with decreased ROS production and restoration of CoQ10 levels in patient fibroblasts co-treated with CoQ10 and riboflavin[141].
Dihydrolipoamide dehydrogenase (E3) deficiency (OMIM# 246900)
The DLD gene encodes dihydrolipoamide dehydrogenase (also known as E3), a common flavoprotein shared by three mitochondrial α-ketoacid dehydrogenase multi-enzyme complexes: pyruvate dehydrogenase complex (PDHC), α-ketoglutarate dehydrogenase complex, and branched-chain α-keto acid dehydrogenase complex. It catalyzes the third stage of the reaction by re-oxidizing the reduced lipoyl moiety of E2 and generating lipoic acid and NADH[142]. E3 is also a component of a fourth mitochondrial multienzyme complex, the glycine cleavage system, where it functions as the L protein, a housekeeping enzyme that, to date, has not appeared to impair the function of the system in vivo when associated with pathogenic variants of DLD[143]. The metabolic derangements incorporate PDHC and TCA cycle defects: increased blood lactate and pyruvate, elevated plasma alanine, BCAA, presence of allo-isoleucine in plasma, and increased urinary lactic, pyruvic, 2-ketoglutaric, and branched-chain 2-hydroxy- and 2-ketoacid[143,144].
The phenotypic spectrum of DLD deficiency includes: (1) classical E3 deficiency, which includes early-onset encephalopathy with progressive hypotonia, failure to thrive, hypoglycemia, ketoacidosis, and encephalopathy or Leigh-like encephalopathy; (2) a primarily hepatic presentation with Reye-like syndrome, wherein patients typically have normal intellect without residual neurologic deficits between acute metabolic episodes; and (3) a milder riboflavin responsive myopathic phenotype with exertional fatigue, intermittent elevation of blood lactate, ketoacidosis, creatine kinase, and mitochondrial proliferation[143,145,146]. Riboflavin supplementation led to complete resolution of muscle weakness, improvement of metabolic abnormalities, partial restoration of the DLD protein, resolution of mitochondrial proliferation in muscle, and reduced ROS production in fibroblasts, supporting the evidence of a chaperone-like effect of riboflavin in promoting DLD protein stability and folding[146].
Ethylmalonic aciduria secondary to ETHE1 dysfunction (OMIM# 602473)
Ethylmalonic encephalopathy (EE) is a rare, devastating, invariably fatal neurodegenerative disease caused by mutations in the ETHE1 gene, which encodes a mitochondrial sulfur dioxygenase critical in hydrogen sulfide (H2S) detoxification[147]. The characteristic clinical features are hydrogen sulfide mediated and include vasculopathy due to diffuse microvasculature injury responsible for multiple necrotic brain lesions, which lead to early-onset psychomotor regression, seizures, and global neurological impairment that subsequently evolve to severe psychomotor delay with spastic tetraparesis. Vascular lesions include diffuse and spontaneous relapsing petechial purpura, orthostatic acrocyanosis, hemorrhagic suffusions of mesothelial surfaces, and intestinal mucosa associated with chronic hemorrhagic diarrhea[148].
Biochemical abnormalities resulting from accumulation of hydrogen sulfide and its derivative thiosulfate in crucial tissues including liver, brain, and colonic mucosa leading to inhibition of both short-chain acyl-CoA dehydrogenase with consequent elevation of ethylmalonate, C4- and C5-acylcarnitines predominantly in muscle and brain, and cytochrome c oxidase deficiency, blocking mitochondrial respiration and increasing lactic acid[147,148]. Urinary thiosulfate is also markedly elevated[149].
Current treatment modalities include the off-label use of common drugs such as N-acetylcysteine and metronidazole to lower the production and promote detoxification of toxic H2S. This combined exposure has produced some encouraging results with amelioration of some neurological abnormalities and marked attenuation or disappearance of the vascular lesions and diarrhea[150], suggesting disease modification from the invariably fatal clinical course of EE[151]. Therapeutic trial with riboflavin and/or Coenzyme Q10 resulted in decreased C4- and C5-acylcarnitines in one patient and mild improvements in chronic mucoid diarrhea and motor and cognitive functions in three patients[152].
AIFM1 deficiency (OMIM#300816, OMIM#310490, OMIM#300614, OMIM#300232)
The X-linked AIFM1 gene encodes the apoptosis-inducing factor (AIF), a mitochondrial flavoprotein with a dual role, first as a FAD-dependent NADH oxidoreductase and second as a caspase-independent death effector which regulates apoptosis[153,154]. AIFM1 mutations have been associated with wide spectrum of clinical phenotypes with X-linked recessive inheritance including a severe, early-onset mitochondrial encephalomyopathy with combined oxidative phosphorylation deficiency[155], prenatal ventriculomegaly[156], the Cowchock syndrome, an X-linked Charcot-Marie-Tooth disease (CMTX4) with axonal sensorimotor neuropathy, deafness and cognitive impairment[157], auditory neuropathy spectrum disorder[158], spondyloepimetaphyseal dysplasia with mental retardation[159], and, more recently, cerebellar ataxia partially responsive to riboflavin[160].
The AIFM1 protein is highly expressed in hairy cells and spiral ganglion within the inner ear, consistent with its crucial role in maintaining normal auditory function[158]. Hence, the predominant phenotypes described with AIFM1 mutations are either mitochondrial encephalomyopathies or sensorineural hearing loss and auditory neuropathy as transmission from the inner ear to the auditory nerve and brain stem is distorted[160]. Cerebellar ataxia, with or without vermian atrophy, was described only recently in relation to AIFM1 mutations, and has further widened the symptomatology[160].
Supplementation with riboflavin was reported to temporarily slow the disease progression in a patient with the severe encephalomyopathy[155]. A clear improvement of ataxia by 44% and 20% was observed in two patients, respectively, as measured by the ICARS score, an objective assessment that quantifies the magnitude of improvement in progressive ataxias, with a score of 5%-20% considered as substantial in providing a clinical impact. Prompt diagnosis through early recognition of constellation of symptoms including ataxia with sensorineural hearing loss especially auditory neuropathy would be important since the symptoms may be partially amenable to riboflavin treatment[160].
Conclusion
Defects of riboflavin metabolism present with a heterogeneous clinical spectrum and variable severity, ranging from severe, early-onset neurodegenerative disorders to adult-onset myopathy. Riboflavin has been used as a potential therapeutic agent in the “mitochondrial cocktail”, particularly in Complex I- and II-related primary mitochondrial disease to ameliorate oxidative stress. There is growing evidence of the beneficial role of riboflavin in treating defects of riboflavin metabolism. Riboflavin supplementation has dramatically led to clinical improvements in riboflavin transporter deficiencies (RTD2 and RTD3), which encompasses BVVL and FL syndromes and mitochondrial FAD transporter deficiency. It has also been proven efficacious in FAD synthase deficiency, late-onset MADD due to ETFDH mutations and deficiencies in ACAD9, dihydrolipoamide dehydrogenase (E3), and AIFM1. In view of the promising therapeutic effects of riboflavin in these disorders, early recognition and treatment including trial of riboflavin supplementation is imperative.
Declarations
Authors’ contributionsMade equal contributions to writing and critically revising the manuscript: Balasubramaniam S, Yaplito-Lee J
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestBoth authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2020.
REFERENCES
1. Lienhart WD, Gudipati V, Macheroux P. The human flavoproteome. Arch Biochem Biophys 2013;535:150-62.
3. Balasubramaniam S, Christodoulou J, Rahman S. Disorders of riboflavin metabolism. J Inherit Metab Dis 2019;42:608-19.
4. Kennedy DO. B Vitamins and the brain: mechanisms, dose and efficacy-a review. Nutrients 2016;8:68.
5. Barile M, Giancaspero TA, Leone P, Galluccio M, Indiveri C. Riboflavin transport and metabolism in humans. J Inherit Metab Dis 2016;39:545-57.
6. Henriques BJ, Rodrigues JV, Gomes CM. Riboflavin and b-oxidation flavoenzymes. In: Preedy VR, editor. B vitamins and folate: chemistry, analysis, function and effects (Food and nutritional components in focus series No. 4). Cambridge: The Royal Society of Chemistry Publishing; 2013. pp. 611-30.
7. Mosegaard S, Dipace G, Bross P, Carlsen J, Gregersen N, et al. Riboflavin Deficiency-Implications for general human health and inborn errors of metabolism. Int J Mol Sci 2020;21:E3847.
8. McCormick DB. Two interconnected B vitamins: riboflavin and pyridoxine. Physiol Rev 1989;69:1170-98.
9. Zempleni J, Galloway JR, McCormick DB. Pharmacokinetics of orally and intravenously administered riboflavin in healthy humans. Am J Clin Nutr 1996;63:54-66.
11. Chastain JL, McCormick DB. Flavin catabolites: identification and quantitation in human urine. Am J Clin Nutr 1987;46:830-4.
12. Suomalainen A, Battersby BJ. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cell Biol 2017;19:77-92.
13. Gorman GS, Chinnery PF, DiMauro S, Hirano M, Kogaet Y, et al. Mitochondrial diseases. Nat Rev Dis Primers 2016;2:16080.
14. Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem 1985;54:1015-69.
15. Van Houten B, Woshner V, Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 2006;5:145-52.
16. Ross WN. Understanding calcium waves and sparks in central neurons. Nat Rev Neurosci 2012;13:157-68.
17. Niyazov DM, Kahler SG, Frye R. Primary mitochondrial disease and secondary mitochondrial dysfunction: importance of distinction for diagnosis and treatment. Mol Syndromol 2016;7:122-37.
18. Yonezawa A, Masura S, Katsura T, Inui K. Identification and functional characterization of a novel human and rat riboflavin transporter, RFT1. Am J Physiol Cell Physiol 2008;295:C632-41.
19. Yonezawa A, Inui K. Novel riboflavin transporter family RFVT/SLC52: identification, nomenclature, functional characterization and genetic diseases of RFVT/SLC52. Mol Aspects Med 2013;34:693-701.
20. Yamamoto S, Inque K, Ohta KY, Fukatsu R, Maeda J, et al. Identification and functional characterization of rat riboflavin transporter 2. J Biochem 2009;145:437-43.
21. Yao Y, Yonezawa A, Yoshimatsu H, Masuda S, Katsura T, et al. Identification and comparative functional characterization of a new human riboflavin transporter hRFT3 expressed in the brain. J Nutr 2010;140:1220-6.
22. Bosch AM, Abeling NG, Ijlst L, Knoester H, van der Pol WL, et al. Brown-Vialetto-Van Laere and Fazio Londe syndrome is associated with a riboflavin transporter defect mimicking mild MADD: a new inborn error of metabolism with potential treatment. J Inherit Metab Dis 2011;34:159-64.
23. Green P, Wiseman M, Crow YJ, Houlden H, Riphagenet S, et al. Brown-Vialetto-Van Laere syndrome, a ponto-bulbar palsy with deafness, is caused by mutations in c20orf54. Am J Hum Genet 2010;86:485-9.
24. Dipti S, Childs AM, Livingston JH, Aggarwal AK, Miller M, et al. Brown-Vialetto-Van Laere syndrome; variability in age at onset and disease progression highlighting the phenotypic overlap with Fazio-Londe disease. Brain Dev 2005;27:443-6.
25. Manole A, Houlden H. Riboflavin transporter deficiency neuronopathy. In: GeneReviews® [Internet]. Seattle (WA): University of Washington; .
26. O’Callaghan B, Bosch AM, Houlden H. An update on the genetics, clinical presentation, and pathomechanisms of human riboflavin transporter deficiency. J Inherit Metab Dis 2019;42:598-607.
27. Chiong MS, Sim KG, Carpenter K, Rhead W, Ho G, et al. Transient multiple acyl-CoA dehydrogenation deficiency in a newborn female caused by maternal riboflavin deficiency. Mol Genet Metab 2007;92:109-14.
28. Ho G, Yonezawa A, Masuda S, Inui K, Sim KG, et al. Maternal riboflavin deficiency, resulting in transient neonatal-onset glutaric aciduria Type 2, is caused by a microdeletion in the riboflavin transporter gene GPR172B. Hum Mutat 2011;32:E1976-84.
29. Mosegaard S, Bruun GH, Flybjerg KF, Bliksrud YT, Gregersen N, et al. An intronic variation in SLC52A1 causes exon skipping and transient riboflavin-responsive multiple acyl-CoA dehydrogenation deficiency. Mol Genet Metab 2017;122:182-8.
30. Schiff M, Veauville-Merllie A, Acquaviva-Bourdain C. SLC25A32 mutations and riboflavin-responsive exercise intolerance. N Engl J Med 2016;374:795-7.
31. Hellebrekers DMEI, Sallevelt SCEH, Theunissen TEJ, Hendrickx ATM, Gottschalk RW, et al. Novel SLC25A32 mutation in a patient with a severe neuromuscular phenotype. Eur J Hum Genet 2017;25:886-8.
32. Santoro V, Kovalenko I, Vriens K, Christen S, Bernthaler A, et al. SLC25A32 sustains cancer cell proliferation by regulating flavin adenine nucleotide (FAD) metabolism. Oncotarget 2020;11:801-12.
33. Giancaspero TA, Colella M, Brizio C, Difonzo G, Fiorino GM, et al. Remaining challenges in cellular flavin cofactor homeostasis and flavoprotein biogenesis. Front Chem 2015;22:30.
34. Olsen RKJ, Konarikova E, Giancaspero TA, Mosegaard S, Boczonadi V, et al. Riboflavin responsive and-non-responsive mutations in FAD synthase cause multiple acyl-CoA dehydrogenase and combined respiratory-chain deficiency. Am J Hum Genet 2016;98:1130-45.
35. Torchetti EM, Brizio C, Colella M, Galluccio M, Giancaspero TA, et al. Mitochondrial localization of human FAD synthetase isoform 1. Mitochondrion 2010;10:263-73.
36. Giancaspero TA, Busco G, Panebianco C, Carmone C, Miccolis A, et al. FAD synthesis and degradation in the nucleus create a local flavin cofactor pool. J Biol Chem 2013;288:29069-80.
37. Leone P, Galluccio M, Barbiroli A, Eberini I, Tolomeo M, et al. Bacterial production, characterization and protein modeling of a novel monofunctional isoform of FAD synthase in humans: an emergency protein? Molecules 2018;23:116-31.
38. Torchetti EM, Bonomi F, Galluccio M, Gianazza E, Giancaspero TA, et al. Human FAD synthase (isoform 2): a component of the machinery that delivers FAD to apo-flavoproteins. FEBS J 2011;278:4434-49.
39. Taylor RW, Pyle A, Griffin H, Blakely EL, Duff J, et al. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA 2014;312:68-77.
40. Auranen MA, Paetau A, Piirilä P, Pohju A, Salmi T, et al. Patient with multiple acyl-CoA dehydrogenation deficiency disease and FLAD1 mutations benefits from riboflavin therapy. Neuromuscul Disord 2017;27:581-4.
41. Yildiz Y, Olsen RKJ, Sivri HS, Akçören Z, Nygaard HH, et al. Post-mortem detection of FLAD1 mutations in 2 Turkish siblings with hypotonia in early infancy. Neuromuscul Disord 2018;28:787-90.
42. Ryder B, Tolomeo M, Nochi Z, Colella M, Barile M, et al. A novel truncating FLAD1 variant, causing multiple Acyl-CoA dehydrogenase deficiency (MADD) in an 8-year-old boy. JIMD Rep 2019;45:37-44.
43. García-Villoria J, De Azua B, Tort F, Mosegaard S, Ugarteburu O, et al. FLAD1, encoding FAD synthase, is mutated in a patient with myopathy, scoliosis and cataracts. Clin Genet 2018;94:592-3.
44. Muru K, Reinson K, Künnapas K, Lilleväli H, Nochi Z, et al. FLAD1-associated multiple acyl-CoA dehydrogenase deficiency identified by newborn screening. Mol Genet Genomic Med 2019;7:e915.
45. Yamada K, Ito M, Kobayashi H, Hasegawa Y, Fukuda S, et al. Flavin adenine dinucleotide synthase deficiency due to FLAD1 mutation presenting as multiple acyl-CoA dehydrogenation deficiency-like disease: a case report. Brain Dev 2019;41:638-42.
46. Karthikeyan S, Zhou Q, Mseeh F, Grishin NV, Osterman AL, et al. Crystal structure of human riboflavin kinase reveals a beta barrel fold and a novel active site arch. Structure 2003;11:265-73.
47. Yazdanpanah B, Wiegmann K, Tchikov V, Krut O, Pongratz C, et al. Riboflavin kinase couples TNF receptor 1 to NADPH oxidase. Nature 2009;460:1159-63.
48. Zhang J, Zhang W, Zou D, Chen G, Wan T, et al. Cloning and functional characterization of ACAD-9, a novel member of human acyl-CoA dehydrogenase family. Biochem. Biophys Res Commun 2002;297:1033-42.
49. Schiff M, Haberberger B, Xia CW, Mohsen AW, Goetzman ES, et al. Complex I assembly function and fatty acid oxidation enzyme activity of ACAD9 both contribute to disease severity in ACAD9 deficiency. Hum Mol Genet 2015;24:3238-47.
50. Nouws J, Te Brinke H, Nijtmans LG, Houten SM. ACAD9, a complex I assembly factor with a moonlighting function in fatty acid oxidation deficiencies. Hum Mol Genet 2014;23:1311-9.
51. Repp BM, Mastantuono E, Alston CL, Schiff M, Haack TB, et al. Clinical, biochemical and genetic spectrum of 70 patients with ACAD9 deficiency: is riboflavin supplementation effective? Orphanet J Rare Dis 2018;13:120.
52. Oey NA, Ruiter JP, Ijlst L, Attie-Bitach T, Vekemans M, et al. Acyl-CoA dehydrogenase 9 (ACAD 9) is the long-chain acyl-CoA dehydrogenase in human embryonic and fetal brain. Biochem Biophys Res Commun ;346:33-7.
53. Lemire BD. Evolution of FOXRED1, an FAD-dependent oxidoreductase necessary for NADH: Ubiquinone oxidoreductase (Complex I) assembly. Biochim. Biophys Acta ;1847:451-7.
54. Lemire BD. Glutathione metabolism links FOXRED1 to NADH: ubiquinone oxidoreductase (complex I) deficiency: a hypothesis. Mitochondrion ;24:105-12.
55. Barbosa-Gouveia S, González-Vioque E, Borges F, Gutiérrez-Solana L, Wintjes L, et al. Identification and characterization of new variants in FOXRED1 gene expands the clinical spectrum associated with mitochondrial complex I deficiency. J Clin Med 2019;8:1262.
56. Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet 2010;42:851-8.
57. Fassone E, Duncan AJ, Taanman JW, Pagnamenta AT, Sadowski MI, et al. FOXRED1, encoding an FAD-dependent oxidoreductase complex-I-specific molecular chaperone, is mutated in infantile-onset mitochondrial encephalopathy. Hum Mol Genet 2010;19:4837-47.
58. Haack TB, Madignier F, Herzer M, Lamantea E, Danhauser K, et al. Mutation screening of 75 candidate genes in 152 complex I deficiency cases identifies pathogenic variants in 16 genes including NDUFB9. J Med Genet 2012;49:83-9.
59. Zurita Rendón O, Antonicka H, Horvath R, Shoubridge EA. A mutation in the flavin adenine dinucleotide-dependent oxidoreductase FOXRED1 results in cell-type-specific assembly defects in oxidative phosphorylation complexes I and II. Mol Cell Biol 2016;36:2132-40.
60. Apatean D, Rakic B, Brunel-Guitton C, Hendson G, Bai R, et al. Congenital lactic acidosis, cerebral cysts and pulmonary hypertension in an infant with FOXRED1 related complex I deficiency. Mol Genet Metab Rep 2019;18:32-8.
61. Formosa LE, Mimaki M, Frazier AE, McKenzie M, Stait TL, et al. Characterization of mitochondrial FOXRED1 in the assembly of respiratory chain complex I. Hum Mol Genet 2015;24:2952-65.
62. Bentinger M, Brismar K, Dallner G. The antioxidant role of coenzyme Q. Mitochondrion 2007;7 Suppl:S41-50.
63. Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Act 2004;1660:171-99.
64. Acosta Lopez MJ, Trevisson E, Canton M, Vazquez-Fonseca L, Morbidoniet V, et al. Vanillic acid restores coenzyme q biosynthesis and ATP production in human cells lacking COQ6. Oxid Med Cell Longe 2019;2019:3904905.
65. Heeringa SF, Chernin G, Chaki M, Zhou W, Sloan AJ, et al. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest 2011;121:2013-24.
66. Cao Q, Li GM, Xu H, Shen Q, Sun L, et al. Coenzyme Q (10) treatment for one child with COQ6 gene mutation induced nephrotic syndrome and literature review. Zhonghua Er Ke Za Zhi 2017;55:135-8. (in Chinese)
67. Gigante M, Diella S, Santangelo L, Trevisson E, Acosta MJ, et al. Further phenotypic heterogeneity of CoQ10 deficiency associated with steroid resistant nephrotic syndrome and novel COQ2 and COQ6 variants. Clin Genet 2017;92:224-6.
68. Park E, Ahn YH, Kang HG, Yoo KH, Won NH, et al. COQ6 mutations in children with steroid-resistant focal segmental glomerulosclerosis and sensorineural hearing loss. Am J Kidney Di 2017;70:139-44.
69. Li GM, Cao Q, Shen Q, Sun L, Zhai YH, et al. Gene mutation analysis in 12 Chinese children with congenital nephrotic syndrome. BMC Nephro 2018;19:382.
70. Stanczyk M, Balasz-Chemielewska I, Lipska-Zietkiewicz B, Tkaczyk M. CoQ10-related sustained remission of proteinuria in a child with COQ6 glomerulopathy-a case report. Pediatr Nephrol 2018;33:2383-7.
71. Yuruk Yildirim Z, Toksoy G, Uyguner O, Nayir A, Yavuz S, et al. Primary coenzyme Q10 Deficiency-6 (COQ10D6): two siblings with variable expressivity of the renal phenotype. Eur J Med Genet 2020;63:103621.
72. Koyun M, Çomak E, Akman S. CoenzymeQ10 therapy in two sisters with CoQ6 mutations with long-term follow-up. Pediatr Nephrol 2019;34:737-8.
73. Ozeir M, Muhlenhoff U, Webert H, Lill R, Fontecave M, et al. Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem Biol 2011;18:1134-42.
74. Ozaltin F. Primary coenzyme Q10 (CoQ 10) deficiencies and related nephropathies. Pediatr Nephrol 2014;29:961-9.
75. Herrmann JM, Riemer J. Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J Biol Chem ;287:4426-33.
76. Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, et al. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol 2007;179:389-95.
77. Di Fonzo A, Ronchi D, Lodi T, Fassone E, Tigano M, et al. The mitochondrial disulfide relay system protein GFER is mutated in autosomal-recessive myopathy with cataract and combined respiratory-chain deficiency. Am J Hum Genet 2009;84:594-604.
78. Ceh-Pavia E, Ang SK, Spiller MP, Lu H. The disease-associated mutation of the mitochondrial thiol oxidase Erv1 impairs cofactor binding during its catalytic reaction. Biochem J 2014;464:449-59.
79. Daithanka VN, Schaefer SA, Dong M, Bahnson BJ, Thorpe C. Structure of the human sulfhydryl oxidase augmenter of liver regeneration and characterization of a human mutation causing an autosomal recessive myopathy. Biochemistry 2010;49:6737-45.
80. Calderwood L, Holm IA, Teot LA, Anselm I. Adrenal insufficiency in mitochondrial disease: a rare case of GFER-related mitochondrial encephalomyopathy and review of the literature. J Child Neurol 2016;31:190-4.
81. Nambot S, Gavrilov D, Thevenon J. Further delineation of a rare recessive encephalomyopathy linked to mutations in GFER thanks to data sharing of whole exome sequencing data. Clin Genet 2017;92:188-98.
82. Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 2014;100:61-77.
83. Hanukoglu I, Jefcoate CR. Mitochondrial cytochrome P-450scc. Mechanism of electron transport by adrenodoxin. J Biol Chem 1980;255:3057-61.
84. Paul A, Drecourt A, Petit F. FDXR mutations cause sensorial neuropathies and expand the spectrum of mitochondrial fe-s-synthesis diseases. Am J Hum Genet 2017;101:630-7.
85. Peng Y, Shinde DN, Valencia AC. Biallelic mutations in the ferredoxin reductase gene cause novel mitochondriopathy with optic atrophy. Human Molecular Genetics 2017;26:4937-50.
86. Slone J, Peng Y, Chamberlin A. Biallelic mutations in FDXR cause neurodegeneration associated with inflammation. J Hum Genet 2018;63:1211-22.
87. Brandt U. Energy converting NADH: quinone oxidoreductase (complex I). Annu Rev Biochem 2006;75:69-92.
88. Scheulke M, Smeitink J, Mariman E, Loeffen J, Plecko B, et al. Mutant NDUFV1 subunit of mitochondrial complex I causes leukodystrophy and myoclonic epilepsy. Nat Genet 1999;21:260-1.
89. Bénit P, Chretien D, Kadhom N, De-Lonlay-Debeney P, Cormier-Daire V, et al. Large-scale deletion and point mutations of the nuclear NDUFV1 and NDUFS1 genes in mitochondrial complex I deficiency. Am J Hum Genet 2001;68:1344-52.
90. Bindu PS, Sonam K, Chiplunkar S. Mitochondrial leukoencephalopathies: a border zone between acquired and inherited white matter disorders in children? Mult Scler Relat Disord 2018;20:84-92.
91. Björkman K, Sofou K, Darin N, Holme E, GKollberg G, et al. Broad phenotypic variability in patients with complex I deficiency due to mutations in NDUFS1 and NDUFV1. Mitochondrion 2015;21:33-40.
92. Breningstall GN, Shoffner J, Patterson RJ. Siblings with leukoencephalopathy. Semin Pediatr Neurol 2008;15:212-5.
93. Bugiani M, Invernizzi F, Alberio S, Briem E, Lamantea E, et al. Clinical and molecular findings in children with complex I deficiency. Biochim Biophys Acta 2004;1659:136-47.
94. Dinwiddie DL, Smith LD, Miller NA, Atherton AM, Farrow EG, et al. Diagnosis of mitochondrial disorders by concomitant next-generation sequencing of the exome and mitochondrial genome. Genomics 2013;102:148-56.
95. Incecik F, Herguner OM, Besen S. Late-onset leigh syndrome due to NDUFV1 mutation in a 10-year-old boy initially presenting with ataxia. J Pediatr Neurosci 2018;13:205-7.
96. Koene S, Rodenburg RJ, Van der Knaap MS, Willemsen MAAP, Sperl W, et al. Natural disease course and genotype-phenotype correlations in Complex I deficiency caused by nuclear gene defects: what we learned from 130 cases. J Inherit Metab Dis 2012;35:737-47.
97. Lal D, Becker K, Motameny S, Altmüller J, Thiele H, et al. Homozygous missense mutation of NDUFV1 as the cause of infantile bilateral striatal necrosis. Neurogenetics 2013;14:85-7.
98. Laugel V, This-Bernd V, Cormier-Daire V, Speeg-Schatz C, de Saint-Martin A, et al. Early-onset ophthalmoplegia in Leigh-like syndrome due to NDUFV1 mutations. Pediatr Neurol 2007;36:54-7.
99. Lee JS, Yoo T, Lee M, Lee Y, Jeon E, et al. Genetic heterogeneity in Leigh syndrome: Highlighting treatable and novel genetic causes. Clin Genet 2020;97:586-94.
100. Lieber DS, Calvo SE, Shanahan K, Slate NG, Liu S, et al. Targeted exome sequencing of suspected mitochondrial disorders. Neurology 2013;80:1762-70.
101. Marin SE, Mesterman R, Robinson B, Rodenburg RJ, Smeitink J, et al. Leigh syndrome associated with mitochondrial complex I deficiency due to novel mutations In NDUFV1 and NDUFS2. Gene 2013;516:162-7.
102. Moran M, Rivera H, Sanchez-Arago M, Blázquez A, Merinero B, et al. Mitochondrial bioenergetics and dynamics interplay in complex I-deficient fibroblasts. Biochim Biophys Acta 2010;1802:443-53.
103. Nafisinia M, Guo Y, Dang X, Li J, Chen Y, et al. Whole exome sequencing identifies the genetic basis of late-onset leigh syndrome in a patient with MRI but little biochemical evidence of a mitochondrial disorder. JIMD Rep 2017;32:117-24.
104. Ortega-Recalde O, Fonseca DJ, Patino LC, Atuesta JJ, Rivera-Nieto C, et al. A novel familial case of diffuse leukodystrophy related to NDUFV1 compound heterozygous mutations. Mitochondrion 2013;13:749-54.
105. Pronicka E, Piekutowska-Abramczuk D, Ciara E, Trubicka J, Rokicki D, et al. New perspective in diagnostics of mitochondrial disorders: two years’ experience with whole-exome sequencing at a national paediatric centre. J Transl Me 2016;14:174.
106. Rubio-Gozalbo ME, Ruitenbeek W, Wendel U, Sengers RC, Trijbels JM, et al. Systemic infantile complex I deficiency with fatal outcome in two brothers. Neuropediatrics 1998;29:43-5.
107. Vilain C, Rens C, Abey A, Balériaux D, Van Bogaert P, et al. A novel NDUFV1 gene mutation in complex I deficiency in consanguineous siblings with brainstem lesions and Leigh syndrome. Clin Genet 2012;82:264-70.
108. Zafeiriou DI, Rodenburg RJ, Scheffer H, van den Heuvel LP, Pouwels PJW, et al. MR spectroscopy and serial magnetic resonance imaging in a patient with mitochondrial cystic leukoencephalopathy due to complex I deficiency and NDUFV1 mutations and mild clinical course. Neuropediatrics 2008;39:172-5.
109. Zhang J, Liu M, Zhang Z, Zhou L, Kong W, et al. Genotypic spectrum and natural history of cavitating leukoencephalopathies in childhood. Pediatr Neurol 2019;94:38-47.
110. Loeffen JL, Smeitink JS, Trijbels JM, Janssen AJ, Triepels RH, et al. Isolated complex I deficiency in children: clinical, biochemical, and genetic aspects. Hum Mutat 2000;15:123-34.
111. Bénit P, Chretien D, Kadhom N, Giurgea I, De Lonlay-Debeney P, et al. Mutant NDUFV2 subunit of mitochondrial complex I causes early onset hypertrophic cardiomyopathy and encephalopathy. Hum Mutat 2003;21:582-6.
112. Pagniez-Mammeri H, Lombes A, Brivet M, Ogier-de Baulny H, Landrieu P, et al. Rapid screening for nuclear genes mutations in isolated respiratory chain complex I defects. Mol Genet Metab 2009;96:196-200.
113. Cameron JM, Mackay N, Feigenbaum A, Tarnopolsky M, Blaseret S, et al. Exome sequencing identifies complex I NDUFV2 mutations as a novel cause of Leigh syndrome. Eur J Paediatr Neurol 2015;19:525-32.
114. Liu HY, Liao PC, Chuang KT, Kao MC. Mitochondrial targeting of human NADH dehydrogenase (ubiquinone) flavoprotein 2 (NDUFV2) and its association with early-onset hypertrophic cardiomyopathy and encephalopathy. J Biomed Sci 2011;18:29.
116. Evenepoel L, Papathomas TG, Krol N, Korpershoek E, de Krijger RR, et al. Toward an improved definition of the genetic and tumor spectrum associated with SDH germ-line mutations. Genet Med 2015;17:610-20.
117. Bausch B, Schiavi F, Ni Y, Welander J, Patocs A, et al. Clinical characterization of the pheochromocytoma and paraganglioma susceptibility genes SDHA, TMEM127, MAX, and SDHAF2 for gene-informed prevention. JAMA Oncol 2017;3:1204-12.
118. Burnichon N, Briere JJ, Libe R, Vescovo L, Rivière J, et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet 2010;19:3011-20.
119. Dwight T, Mann K, Benn DE, Robinson BG, McKelvie P, et al. Familial SDHA mutation associated with pituitary adenoma and pheochromocytoma/paraganglioma. J Clin Endocrinol Metab 2013;98:E1103-8.
121. Ozluk Y, Taheri D, Matosi A, Sanli O, Berker NK, et al. Renal carcinoma associated with a novel succinate dehydrogenase A mutation: a case report and review of literature of a rare subtype of renal carcinoma. Hum Pathol 2015;46:1951-5.
122. Burgeois M, Goutieres F, Chretien D, Rustin P, Munnich A, et al. Deficiency in complex II of the respiratory chain, presenting as a leukodystrophy in two sisters with Leigh syndrome. Brain Dev 1992;14:404-8.
123. Bourgeron T, Rustin P, Chretien D, Birch-Machin M, Bourgeois M, et al. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat Genet 1995;11:144-9.
124. Alston CL, Davison JE, Meloni F, van der Westhuizen FH, He L, et al. Recessive germline SDHA and SDHB mutations causing leukodystrophy and isolated mitochondrial complex II deficiency. J Med Genet 2012;49:569-77.
125. Helman G, Caldovic L, Whitehead MT, Simons C, Brockmann K, et al. Magnetic resonance imaging spectrum of succinate dehydrogenase-related infantile leukoencephalopathy. Ann Neurol 2016;79:379-86.
126. Horvath R, Abicht A, Holinski-Feder E, Laner A, Gempel K, et al. Leigh syndrome caused by mutations in the flavoprotein (Fp) subunit of succinate dehydrogenase (SDHA). J Neurol Neurosurg Psychiatry 2006;77:74-6.
127. Ma YY, Wu TF, Liu YP, Wang Q, Li XY, et al. Two compound frame-shift mutations in succinate dehydrogenase gene of a Chinese boy with encephalopathy. Brain Dev 2014;36:394-8.
128. Renkema GH, Wortmann SB, Smeets RJ, Venselaar H, Antoine M, et al. SDHA mutations causing a multisystem mitochondrial disease: novel mutations and genetic overlap with hereditary tumors. Eur J Hum Genet 2015;23:202-9.
129. Van Coster R, Seneca S, Smet J, Van Hecke R, Gerlo E, et al. Homozygous Gly555Glu mutation in the nuclear encoded 70 kDa flavoprotein gene causes instability of the respiratory chain complex II. Am J Med Genet A 2003;120A:13-8.
130. Pagnamenta AT, Hargreaves IP, Duncan AJ, Taanman JW, Heales SJ, et al. Phenotypic variability of mitochondrial disease caused by a nuclear mutation in complex II. Mol Genet Metab 2006;89:214-21.
131. Parfait B, Chretein D, Rotig A, Marsac C, Munnich A, et al. Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum Genet 2000;106:236-43.
132. Levitas A, Muhammad E, Harel G, Saada A, Caspi VC, et al. Familial neonatal isolated cardiomyopathy caused by a mutation in the flavoprotein subunit of succinate dehydrogenase. Eur J Hum Genet 2010;18:1160-5.
133. Taylor RW, Birch-Machin MA, Schaefer J, Taylor L, Shakir R, et al. Deficiency of complex II of the mitochondrial respiratory chain in late-onset optic atrophy and ataxia. Ann Neurol 1996;39:224-32.
134. Birch-Machin MA, Taylor RW, Cochran B, Ackrell BA, Turnbull DM. Late-onset optic atrophy, ataxia, and myopathy associated with a mutation of a complex II gene. Ann Neurol 2000;48:330-5.
135. Courage C, Jackson CB, Hahn D, Euro L, Nuoffer JM, et al. SDHA mutation with dominant transmission results in complex II deficiency with ocular, cardiac, and neurologic involvement. Am J Med Genet A 2017;173:225-30.
136. Olsen RK, Andresen BS, Christensen E, Bross P, Skovby F, et al. Clear relationship between ETF/ETFDH genotype and phenotype in patients with multiple acyl-CoA dehydrogenation deficiency. Hum Mutat 2003;22:12-23.
137. Frerman FE, Goodman SI. The Metabolic and Molecular Bases of Inherited Disease. In: Scriver CR, Sly WS, Childs B, Beaudet AL, Valle D, Kinzler KW, Vogelstein B, editors. 2001. Defects of Electron Transfer Flavoprotein and Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase: Glutaric Acidemia Type II. Available from: https://ommbid.mhmedical.com/content.aspx?bookid=2709§ionid=225088261. [Last accessed on 27 Jul 2020].
138. Grünert SC. Clinical and genetical heterogeneity of late-onset multiple acyl-coenzyme dehydrogenase deficiency. Orphanet J Rare Dis 2014;9:117.
139. Morris AAM, Spiekerkoetter U. Disorders of mitochondrial fatty acid oxidation and Related Metabolic Pathways. In: Saudubray JM, van den Berghe G, Walter JH, editors. Inborn metabolic diseases: diagnosis and treatment. Berlin: Springer; 2016.
140. Horvath R. Update on clinical aspects and treatment of selected vitamin-responsive disorders II (riboflavin and CoQ10). J Inherit Metab Dis 2012;35:679-87.
141. Cornelius N, Corydon TJ, Gregersen N, Olsen RKJ. Cellular consequences of oxidative stress in riboflavin responsive multiple acyl-CoA dehydrogenation deficiency patient fibroblasts. Hum Mol Genet 2014;23:4285-301.
142. Cameron JM, Levandovskiy V, Mackay N, Raiman J, Renaud DL, et al. Novel mutations in dihydrolipoamide dehydrogenase deficiency in two cousins with borderline normal PDH complex activity. Am J Med Genet A 2015;140:1542-52.
143. Quinonez SC, Thoene JG. Dihydrolipoamide dehydrogenase deficiency. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews® [Internet]. 2014. Seattle (WA): University of Washington, Seattle; 1993-2020. Available from: https://www.ncbi.nlm.nih.gov/books/NBK220444/. [Last accessed on 27 Jul 2020].
144. De Meirleir LJ, Garcia-Cazorla A, Brivet M. Disorders of pyruvate metabolism and the tricarboxylic acid cycle. Inborn metabolic diseases: diagnosis and treatment. In: Saudubray JM, Baumgartner MR, Walter J, editors. Berlin: Springer; 2016. pp. 161-74.
145. Quintana E, Pineda M, Font A, Vilaseca MA, Tort F, et al. Dihydrolipoamide dehydrogenase (DLD) deficiency in a Spanish patient with myopathic presentation due to a new mutation in the interface domain. J Inherit Metab Dis 2010;33:S315-9.
146. Carrozzo R, Torraco A, Fiermonte G, Martinelli D, Nottia MD, et al. Riboflavin responsive mitochondrial myopathy is a new phenotype of dihydrolipoamide dehydrogenase deficiency. The chaperon-like effect of vitamin B2. Mitochondrion 2014;18:49-57.
147. Tiranti V, Viscomi C, Hildebrandt T, Di Meo I, Mineri R, et al. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat Med 2009;15:200-5.
148. Di Meo I, Lamperti C, Tiranti V. Ethylmalonic encephalopathy. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews [Internet] 2017. Seattle (WA): University of Washington; .
149. Kožich V, Ditrói T, Sokolová J, Křížková M, Krijt J, et al. Metabolism of sulfur compounds in homocystinurias. Br J Pharmacol 2019;176:594-606.
150. Viscomi C, Burlina AB, Dweikat I, Savoiardo M, Lamperti C, et al. Combined treatment with oral metronidazole and N-ace-tylcysteine is effective in ethylmalonic encephalopathy. Nat Med 2010;16:869-71.
151. Di Meo I, Lamperti C, Tiranti V. Mitochondrial diseases caused by toxic compound accumulation: from etiopathology to therapeutic approaches. EMBO Mol Med 2015;7:1257-66.
152. Yoon HR, Hahn SH, Ahn YM, Jang SH, Shin YJ, et al. Therapeutic trial in the first three Asian cases of ethylmalonic encephalopathy: response to riboflavin. J Inherit Metab Dis 2001;24:870-3.
153. Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 1999;397:441e6.
154. Joza N, Pospisilik JA, Hangen E, Hanada T, Modjtahedi N, et al. AIF: not just an apoptosis-inducing factor. Ann NY Acad Sci 2009;1171:2-11.
155. Ghezzi D, Sevrioukova I, Invernizzi F, Lamperti C, Mora M, et al. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am J Hum Genet 2010;86:639-49.
156. Berger I, Ben-Neriah Z, Dor-Wolman T, Shaag A, Saada A, et al. Early prenatal ventriculomegaly due to an aifm1 mutation identified by linkage analysis and whole exome sequencing. Mol Genet Metab 2011;104:517-20.
157. Rinaldi C, Grunseich C, Sevrioukova IF, Schindler A, Horkayne-Szakaly I, et al. Cowchock syndrome is associated with a mutation in apoptosis-inducing factor. Am J Hum Genet 2012;91:1095-102.
158. Zong L, Guan J, Ealy M, Zhang Q, Wang D, et al. Mutations in apoptosis-inducing factor cause x-linked recessive auditory neuropathy spectrum disorder. J Med Genet 2015;52:523-31.
159. Mierzewska H, Rydzanicz M, Bieganski T, Kosinska J, Mierzewska-Schmidt M, et al. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with aifm1 mutation - a novel phenotype of the mitochondrial disease. Clin Genet 2017;91:30-7.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Balasubramaniam S, Yaplito-Lee J. Riboflavin metabolism: role in mitochondrial function. J Transl Genet Genom 2020;4:285-306. http://dx.doi.org/10.20517/jtgg.2020.34
AMA Style
Balasubramaniam S, Yaplito-Lee J. Riboflavin metabolism: role in mitochondrial function. Journal of Translational Genetics and Genomics. 2020; 4(4): 285-306. http://dx.doi.org/10.20517/jtgg.2020.34
Chicago/Turabian Style
Balasubramaniam, Shanti, Joy Yaplito-Lee. 2020. "Riboflavin metabolism: role in mitochondrial function" Journal of Translational Genetics and Genomics. 4, no.4: 285-306. http://dx.doi.org/10.20517/jtgg.2020.34
ACS Style
Balasubramaniam, S.; Yaplito-Lee J. Riboflavin metabolism: role in mitochondrial function. J. Transl. Genet. Genom. 2020, 4, 285-306. http://dx.doi.org/10.20517/jtgg.2020.34
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 121 clicks
Cite This Article 121 clicks


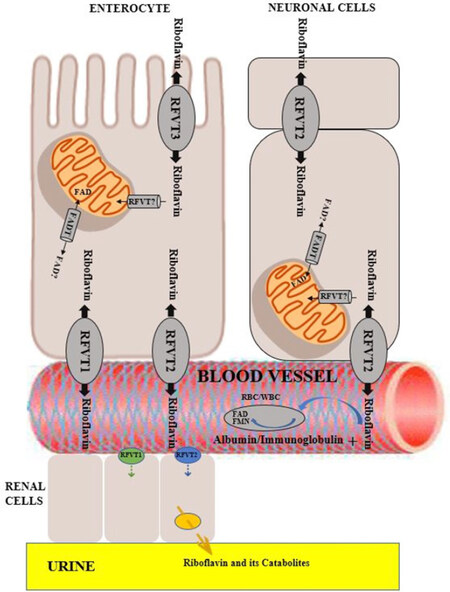
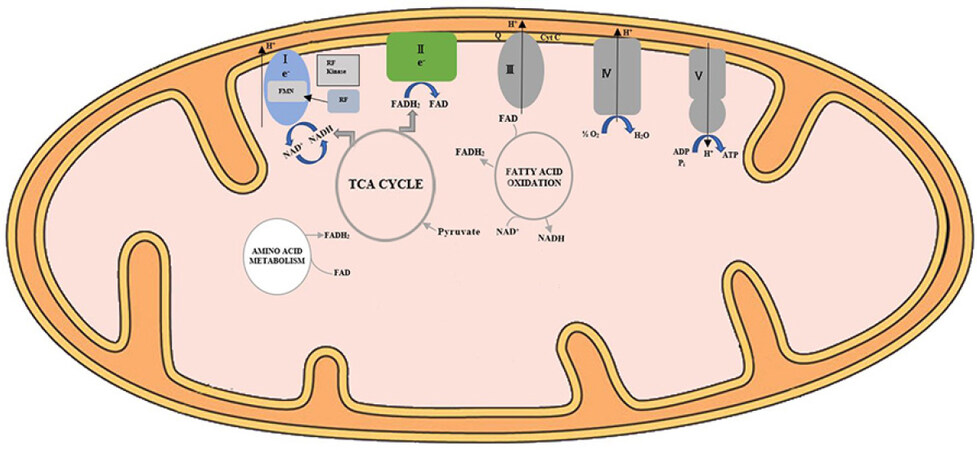









Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.