Roles of interacting stress-related genes in lifespan regulation: insights for translating experimental findings to humans
Abstract
Aim: Experimental studies provided numerous evidence that caloric/dietary restriction may improve health and increase the lifespan of laboratory animals, and that the interplay among molecules that sense cellular stress signals and those regulating cell survival can play a crucial role in cell response to nutritional stressors. However, it is unclear whether the interplay among corresponding genes also plays a role in human health and lifespan.
Methods: Literature about roles of cellular stressors have been reviewed, such as amino acid deprivation, and the integrated stress response (ISR) pathway in health and aging. Single nucleotide polymorphisms (SNPs) in two candidate genes (GCN2/EIF2AK4 and CHOP/DDI3T) that are closely involved in the cellular stress response to amino acid starvation, have been selected using information from experimental studies. Associations of these SNPs and their interactions with human survival in the Health and Retirement Study data have been estimated. The impact of collective associations of multiple interacting SNP pairs on survival has been evaluated, using a recently developed composite index: the SNP-specific Interaction Polygenic Risk Score (SIPRS).
Results: Significant interactions have been found between SNPs from GCN2/EIF2AK4 and CHOP/DDI3T genes that were associated with survival 85+ compared to survival between ages 75 and 85 in the total sample (males and females combined) and in females only. This may reflect sex differences in genetic regulation of the human lifespan. Highly statistically significant associations of SIPRS [constructed for the rs16970024 (GCN2/EIF2AK4) and rs697221 (CHOP/DDIT3)] with survival in both sexes also been found in this study.
Conclusion: Identifying associations of the genetic interactions with human survival is an important step in translating the knowledge from experimental to human aging research. Significant associations of multiple SNPxSNP interactions in ISR genes with survival to the oldest old age that have been found in this study, can help uncover mechanisms of multifactorial regulation of human lifespan and its heterogeneity.
Keywords
INTRODUCTION
The multifactorial nature of aging, health, and lifespan-related traits is broadly recognized but understudied
It is generally acknowledged that human lifespan, aging, and age-associated health disorders are multifactorial traits resulting from the complex interplay among numerous genetic and non-genetic factors. Observed correlations between biomarkers of biological aging and age-associated diseases indicate a possibility of improving health and increasing lifespan through deceleration of the aging-related processes in the body. A better understanding of the mechanisms of multifactorial regulation of respective traits could substantially facilitate the realization of this idea.
Numerous experiments using animal models were performed to improve such understanding. Surprisingly, a number of studies revealed that mutations in just one gene in C. elegans could substantially increase the lifespan of laboratory animals (reviewed in[1]). E.g., Johnson et al.[2,3] identified the long-lived mutant of C. elegans called age-1. Later, other mutants with substantially longer survival compared to wild animals were detected[1,4,5]. Note that the effects of such mutations on lifespan in other species are much less pronounced.
Many other experimental studies discovered that better health and longevity could be achieved in animals from different species exposed to caloric/dietary restriction (CR/DR) (reviewed in[6-9]). Further research showed that separate components of the diet, including carbohydrates[10,11], lipids[12], proteins[13-16], vitamins[17], minerals[18], fiber[19], and water, can influence aging and lifespan through different albeit often interacting genetic regulatory mechanisms. Experiments with different mutants of C. elegans[20] and different strains of mice[21] overall suggested that the effects of CR/DR on lifespan can have a strong genetic component. The genetic mechanisms involved in lifespan regulation in response to amino acid deprivation were linked to genes from the mTOR signaling pathway[22,23]. It has also been shown that amino acid deprivation may influence lifespan by activating interplay among genes from the integrated stress response (ISR) pathway with the GCN2/EIF2AK4 gene serving as a sensor of such stress signals and the CHOP/DDIT3 gene serving as a regulator of cell’s fate deciding between autophagy, cell cycle arrest, or apoptosis[24-26]. Apoptosis is a cellular choice when stress response cannot restore the normal cell’s functioning.
The genetic epidemiological genome-wide and candidate genes single locus studies of human aging, health, and longevity traits made substantial progress in identifying genetic variants associated with these traits. Surprisingly, genes from many signaling and metabolic pathways whose roles in such traits were well established in experimental studies did not show statistically significant associations with these traits. This lack of consistency between results of genetic association studies of human data and information from experimental studies might be explained by the fact that biological mechanisms of complex traits regulation result from a complicated interplay between many genetic and non-genetic factors that may differ in humans and laboratory animals. Studying such interplay requires more sophisticated approaches than used in traditional genetic association studies that test the association of one SNP at a time with a given trait. It means that genetic association studies of multifactorial traits should include analysis of genetic interactions and collective actions of interactions of many genetic and non-genetic factors. To address this problem, many statistical approaches that aimed to detect associations of gene-gene (GxG) interactions have been proposed during the last decades.
Critical reviews of the methods, related software packages used to detect the interactions between genetic loci that contribute to human genetic diseases and the difficulties in determining the biological relevance of statistical interactions are provided in the papers[27-30]. Most recent reviews describe various extensions of the Multifactor Dimensionality Reduction (MDR) approach[31-34], using entropy in genetic interaction analyses[35,36], implementation of machine learning techniques to study epistasis[28,37,38], as well as many other approaches that differ in definitions of genetic interaction, the accuracy of calculations, and in computation time[39-45].
This paper used the INTERSNP software package[46] that implements a logistic regression framework. This approach allows for evaluating associations of genetic interactions with complex traits in the presence of observed covariates. It has been successfully used in our earlier genetic analyses of genetic interactions[47]. Useful information about other methods can also be found in the review paper[48].
Information about genes involved in multifactorial regulation of aging, health, and lifespan-related traits in laboratory animals serves as a source of useful insights concerning genetic mechanisms that might regulate these traits in humans. This information is used for selecting candidate medications appropriate for testing in clinical trials. The failure of many expensive clinical trials to identify a proper medication [e.g., in case of Alzheimer’s disease (AD)] indicates the need to find more reliable and less expensive ways of testing whether genetic connections detected in experimental studies exist in humans. It is proposed that such testing can be done by applying genetic epidemiological methods to available human data on genotyped individuals collected in human longitudinal and cross-sectional studies. The efficient analysis of such data requires a convenient conceptual framework that would allow the researchers to perform comprehensive analyses of biological mechanisms and efficiently integrate research findings.
Stress-related conceptual framework allows for linking together stressors, sensors of stress signals, and genes from stress response pathways as key players in mechanisms of multifactorial regulation of complex traits.
The use of a stress-related conceptual framework might be beneficial for studying multifactorial regulation of complex traits because it allows for selecting and linking together non-genetic factors (e.g., associated with cellular stressors) and genes involved in cellular stress response (e.g., such as ISR). It has been recently shown that interplay between the GCN2/EIF2AK4 gene that serves as one of the sensors of cellular stress signals in the ISR pathway and the gene CHOP/DDIT3 involved in the regulation of autophagy and apoptosis may play a crucial role in the regulation of aging, health, and lifespan/survival traits in laboratory animals[25,49]. This observation allows us to hypothesize that interplay between these genes may influence these traits in humans. Testing this hypothesis using human data would be an important step forward in the translation of knowledge from experimental studies to humans. This is because the interplay between the GCN2/EIF2AK4 and CHOP/DDIT3 genes influences aging, health, and lifespan/survival traits in laboratory animals[25,50,51] does not mean that these genes play the same roles in humans.
Does the interplay between the GCN2/EIF2AK4 and CHOP/DDIT3 genes influence human lifespan?
This paper reviewed experimental evidence about genetic mechanisms that regulate the effects of cellular stress on aging, health, and lifespan/survival traits. These effects are manifested at the cellular level and involve genes from the ISR pathway. To illustrate our approach to the analysis of the effects of genetic interplay on these traits, two genes have been selected from the ISR pathway. One, the GCN2/EIF2AK4 gene, becomes activated by several cellular stressors, including amino acid starvation (deprivation). The transformed signal sent from this gene (but not the initial stress signal) activates other genes in the ISR pathway, including the CHOP/DDIT3 gene, which is the second gene selected for analysis. The product of this gene, among other things, influences the cells’ fate: when stress is mild, and the duration of the stress response is relatively short, the cell has high chances to survive; alternatively, under strong or persistent stress, the CHOP/DDIT3 gene is more likely to activate the process of apoptosis for this cell. The strength and the duration of cellular stress response to a large extent might be determined by the GCN2/EIF2AK4 gene polymorphisms. The cells’ fate has important consequences for the organism’s health and survival outcomes. Because of the importance of stress-related conceptual framework used in our analysis, a brief description of ISR is given below.
Rationale for selecting the GCN2/EIF2AK4 and CHOP/DDIT3 genes involved in the ISR pathway
In response to various stressors disturbing normal cellular functioning, eukaryotic cells activate an evolutionary conserved adaptive machinery - the ISR[49]. Depending on the strength and duration of the stress response, ISR determines the fate of the cell[52]. Cellular stressors may be intrinsic (e.g., misfolded proteins, genetic polymorphisms[53]) or external (e.g., nutrient deprivation, viral infection, hypoxia, UV-irradiation, and others[54-56]). Experimental and clinical studies provide evidence about involvement of ISR in lifespan regulation[57,58], as well as in the development of aging-related diseases including cognitive and neurodegenerative disorders[49,55], cancer[59-61], pulmonary disease[62], atherosclerosis[63,64], diabetes[65] and other metabolic disorders[66].
ISR responds to cellular stressors by changing the process of protein synthesis[49,52]. This process starts with the phosphorylation of eukaryotic translation initiation factor 2 alpha (EIF2A) by one of the four members of the EIF2A kinase family, which sense cellular stress signals. These include a heme-regulated inhibitor kinase (HRI/EIF2AK1), an interferon-induced, double-stranded RNA (dsRNA)-activated protein kinase (PKR/EIF2AK2), a protein kinase R (PKR)-like endoplasmic reticulum (ER) kinase (PERK/EIF2AK3), and a general control nonderepressible 2 kinase (GCN2/EIF2AK4). Additional details about these kinases are described below.
Heme regulating inhibitor kinase (HRI/EIF2AK1) is an enzyme that in humans is encoded by the EIF2AK1 gene. Heme is an iron-containing compound that forms the non-protein part of hemoglobin, the substance inside red blood cells that binds to oxygen in the lungs and carries it to the tissues. HRI/EIF2AK1 is an intracellular heme sensor that coordinates heme and globin synthesis in erythropoiesis by inhibiting protein synthesis of globin and heme biosynthetic enzymes during heme deficiency. HRI is also activated by arsenite-induced oxidative stress, heat shock, nitric oxide, 26S proteasome inhibition, and osmotic stress. These types of stressors activate HRI independently of heme but require the presence of heat shock proteins HSP90 and HSP70. Denatured proteins and oxidative stress also activate HRI[56,67,68].
Protein kinase R (PKR/EIF2AK2) is an enzyme that in humans is encoded by the EIF2AK2 gene. In addition to dsRNA that can be introduced to the cell by a viral infection, PKR is also activated by oxidative and ER stress, growth factor deprivation, cytokines, bacterial infections, ribotoxic stress[69], caspase activity in the early stages of apoptosis[70]. It can also be activated by the protein PACT (that in humans is encoded by the PRKRA gene) or by heparin and other cellular stress signals[71-73].
Protein kinase R-like endoplasmic reticulum kinase (PERK/EF2AK3) is an enzyme that in humans is encoded by the EIF2AK3 gene. PERK is activated by accumulation of misfolded (unfolded) proteins in the ER, perturbations in calcium homeostasis, cellular energy, mitochondrial stress (including uncoupling), or redox status[74,75]. It has also been reported to respond to ATP depletion and subsequent sarcoplasmic/ER Ca2+-ATPase pump inhibition in the context of glucose deprivation in neuronal cells and pancreatic β cells[76-78]. It initiates the unfolded protein response[79,80]. PERK plays an important role in Alzheimer’s and other neurodegenerative diseases[81-83].
GCN2/EIF2AK4 is an enzyme that in humans is encoded by the EIF2AK4 gene. GCN2 is evolutionarily conserved from yeasts to humans and plays a key role in modulating amino acid metabolism. It is activated in response to amino acid deprivation when it binds to deacylated transfer RNAs (tRNAs) via histidyl-tRNA synthetase-related domain[25,84-87]. GCN2 can also be activated by other stressors, including ultraviolet irradiation, viral infection, serum starvation, glucose deprivation, and oxidative stress. Recent work shows that GCN2 strongly activates by binding to ribosomal protein, suggesting that GCN2 actively monitors mRNA translation[25,84-87]. Recently, a pivotal role for GCN2 in response to membrane damage has been uncovered[88,89]. Finally, GCN2, a crucial regulator of amino acid metabolism, is necessary for the metabolic homeostasis of tumor cells. Tumors lacking GCN2 or ATF4 grow more slowly[61]. Thus, in cancers where amino acids are scarce, targeting the GCN2 branch of the ISR may be beneficial. Indeed, combination treatment with L-asparaginase and GCN2 inhibitors causes apoptosis in several cancer cell types[49]. GCN2 upregulates a coordinately expressed set of genes involved in amino acid biosynthesis and metabolism[90].
The EIF2A phosphorylation by one of four kinases results in a decrease in global protein synthesis and the enhancing translation of the activating transcription factor ATF4 and several other genes acting together to restore cellular homeostasis[91,92]. ATF4 mediates the induction of ATF3 and GADD34/PPP1R15A, which dephosphorylates EIF2A-P and leads to the termination of ISR[93,94]. Chronic ISR activates CHOP/DDIT3 leading to apoptosis[60,95]. EIF2A phosphorylation also blocks the action of EIF2B, resulting in a general reduction in protein synthesis and the upregulation of selected genes. One of such genes is the transcription factor ATF4. The product of this gene plays a critical role in the regulation of obesity, glucose homeostasis, energy expenditure, and neural plasticity[96,97].
Under stress conditions, increased ATF4 expression can activate several transcriptional programs that will ultimately determine the cell fate-from re-establishment of homeostasis to cell death[98]. The ability of ATF4 to interact with multiple other transcription factors makes its target genes highly dependent on stress intensity and cellular context[61,87,99-101]. For example, when acting in combination with ATF3, ATF4 contributes to re-establishing cellular homeostasis and survival promotion[102]. Conversely, when interacting with CHOP, ATF4 promotes cell death[93]. In addition to the interacting partners that cooperate with ATF4 to promote transcription of target genes, another set of interacting partners prevent ATF4 transcriptional activity, as is the case for PHD3 during hypoxia[96,97] and TRIB3 during amino acid starvation[103].
METHODS
Data
This paper aims to show how information from experimental data can be used for testing the connection between genetic factors and survival traits in humans. For this, a set of data is needed on genotyped individuals with large sample size. The Health and Retirement Study (HRS) data on white individuals satisfies this requirement. The information on white HRS study subjects is shown in Table 1.
Summary statistics for white HRS respondents with genetic data
| Males | Females | Total | ||||
| Case | Control | Case | Control | Case | Control | |
| N | 1056 | 1592 | 1508 | 1986 | 2564 | 3578 |
| Dichotomous variables: N (Percentage) | ||||||
| Education, Highschool+ | 440 (91.06) | 536 (94.07) | 803 (94.29) | 818 (96.91) | 1243 (92.95) | 1354 (95.63) |
| Ever smoked | 703 (67.60) | 845 (70.53) | 615 (41.84) | 759 (52.13) | 1318(52.51) | 1604 (60.44) |
The genetic data - 2.5 million single nucleotide polymorphisms (SNPs) - were produced on the Illumina platform using Illumina’s Human Omni2.5-Quad (Omni2.5) BeadChip methodology on 15,620 individuals (6472 males and 9148 females).
Quality control (QC) was performed before running the analysis using two procedures. The first, based on the protocol proposed in[104], resulted in: 129 individuals dropped because of failure in the check of duplicates, missingness rate (5%), heterozygosity outlier (± 3 SD), sex mismatch, and divergent ancestry outlier (± 8 SD); 538,451 variants were removed due to minor allele frequency < 1%, 69,947 variants were removed due to genotyping missing rate higher than 5%; and 411,945 variants were removed based on Hardy-Weinberg test (HWE, P-value < 1.0E-7). The second procedure used the protocol proposed in[105]. Individuals and SNP which passed either the first QC step or the second step remained in the final dataset. This procedure allowed us to increase the number of study subjects used in genetic analysis. It led to 15,492 individuals and 1,267,439 variants cleaned and mapped to the human reference genome GRCh38 for further analysis.
The following dichotomous survival trait (ST) was used in genetic interaction analysis: case: LS ≥ 85; control: 75 ≤ LS < 85 (or age at last follow-up) because we were especially interested in the effects of genetic factors on survival at ages 85+ compared with that around age 80 (± 5 years). Our earlier studies[106,107] suggested that the age around 80 might be a "switching point" in the course of aging, characterized by declining or leveling-off (after a prior increase) risks of some major diseases (e.g., cancers, asthma, CVD, diabetes). Such behavior of risk trajectories could be due to selection, under-diagnosis, or the aging itself, so that some aging-related changes in the body would negatively affect health and survival chances before age 80 but become protective afterward[107]. To estimate the association of interacting SNP pairs with ST, the logistic regression model as implemented in INTERSNP software was used (with the interaction term being the quantity of interest)[46]. Education, smoking status, sex, and first five principal components (PC1-PC5) were included as observed covariates.
Evaluating collective association of interactive SNP pairs with the survival trait
Because of the multifactorial nature of age-related health and lifespan-related traits, the interactions of SNPs from the GCN2/EIF2AK4 or CHOP/DDIT3 genes with SNPs from many other genes may also contribute to these traits. The interacting SNP pairs associated with these traits can be detected in the genome-wide-like association studies of interacting SNP pairs in which one SNP from the GCN2/EIF2AK4 or CHOP/DDIT3 genes is fixed, and others are all SNPs available for a given dataset that passed the quality control procedure. The results of this analysis can be used for constructing a SNP-specific composite index that can measure the association of many interacting SNP pairs with health and lifespan-related traits. This measure extends the notion of polygenic risk score (PRS), widely used in the genetic epidemiological studies of age-related diseases and longevity[108-116]. A simple measure of such capacity for a given SNP* might be the number of detected associations of interacting SNP pairs (with a fixed SNP*) whose P-values did not exceed a given P-value threshold (similar to the genetic dose index described in ref[113]). This paper introduced a measure of interactive capacity of a given SNP* involved in the lifespan regulation called “SNP-specific Interaction Polygenic Risk Score” (SIPRS). The construction of such indices and some of their properties related to multifactorial regulation of AD are described in Yashin et al.[47].
RESULTS
The number of pairs of interacting SNPs in the two genes
To evaluate the significance of associations of the SNPxSNP interactions with the survival trait, the number of tested SNP pairs is needed to know when deciding about the presence or absence of the association of the interacting SNP pair with the trait. This number depends on the population under study and the genotyping platform. In the HRS data, after the quality control procedure, the GCN2/EIF2AK4 and the CHOP/DDIT3 genes have 63 and 8 SNPs, respectively. The analysis of associations of these SNPs with the survival trait (ST) (defined in the Data and methods section) showed that the associations of each of the 71 SNPs with survival trait did not reach the nominal level (P ≤ 0.05) of statistical significance. This allows for the hypothesis that the contribution of these genes to survival might be realized through their interaction effects.
To evaluate associations of interaction between the GCN2/EIF2AK4 and CHOP/DDIT3 genes with survival traits, a set of 504 potentially interacting SNP pairs has been used to estimate the set of probabilities of the type I error, which would happen if the decision about the presence of association of each SNP pair with survival trait would be made. Note that this procedure did not involve the “decision making” about the presence or absence of association of the SNPxSNP interaction with the trait. Probabilities of type I error have been calculated for each SNP pair and, hence, no correction for multiple testing is needed at this stage. Note that the Bonferroni correction for multiple testing, in this case, would be 9.92E-05.
Significant associations of interacting SNP pairs between the GCN2/EIF2AK4 and CHOP/DDIT3 genes with human survival trait
The presence of linkage disequilibrium (LD) between SNPs related to each of two genes allows for reducing the number of potentially interacting SNP pairs tested for their associations with lifespan and for increasing the Bonferroni correction threshold. For this, the LD regions in each gene and selected one SNP pair as a representative for each such region have been identified. In the analysis, the SNP pair representatives are those who have the smallest value of the type I error among SNP pairs in this region. This clumping procedure used R2 = 0.1 LD threshold and resulted in 8 independent SNP pairs: 8 independent SNPs from the GCN2/EIF2AK4 gene and 1 independent SNP from the CHOP/DDIT3 gene. It gives us a set of 8 SNP pairs that can now be used for testing the null hypothesis about the absence of association of the interacting SNP pairs with survival traits. The smallest P-value resulted from the analysis of the association of SNPxSNP interaction with survival trait is 3.80E-03 for SNP rs16970024 from the GCN2/EIF2AK4 gene interacting with SNP rs697221 related to the CHOP/DDIT3 gene [Table 2]. All other interactions have P-values exceeding 1.00E-03. With eight SNP pairs tested in this analysis, the Bonferroni correction provides us with the P-value threshold 6.25E-03 for testing the null hypothesis, which is larger than 3.80E-03. These results are summarized in Table 2.
Associations of interactions between SNPs in GCN2/EIF2AK4 and CHOP/DDIT3 genes with *survival trait after clumping with threshold R2 = 0.1
| rsid1 | 1EA/NEA | 1MA | p1 | 1MAF | rsid2 | 2EA/NEA | 2MA | p2 | 2MAF | b12 | p12 |
| rs16970024 | A/G | G | 0.81 | 0.06 | rs697221 | G/A | A | 0.98 | 0.14 | 0.60 | 3.88E-03 |
| rs72731410 | G/A | A | 0.97 | 0.03 | rs697221 | G/A | A | 0.98 | 0.14 | 0.75 | 2.81E-02 |
| rs3736290 | A/C | C | 0.08 | 0.47 | rs697221 | G/A | A | 0.98 | 0.14 | 0.19 | 5.20E-02 |
| rs7169266 | A/G | G | 0,60 | 0.03 | rs697221 | G/A | A | 0.98 | 0.14 | -0.40 | 1.13E-01 |
| rs76182620 | A/G | G | 0.54 | 0.05 | rs697221 | G/A | A | 0.98 | 0.14 | -0.34 | 1.35E-01 |
| rs12442731 | A/G | G | 0.17 | 0.44 | rs697221 | G/A | A | 0.98 | 0.14 | -0.14 | 1.87E-01 |
| rs117584784 | G/A | A | 0.44 | 0.02 | rs697221 | G/A | A | 0.98 | 0.14 | -0.21 | 4.82E-01 |
| rs566792 | G/A | A | 069 | 0.13 | rs697221 | G/A | A | 0.98 | 0.14 | -0.05 | 0.731 |
Separate analysis of males and females revealed a significant association of interactions between the SNPs rs16970024 and rs697221 with female survival, with a P-value of 3.80E-03. No significant associations of the interacting SNPs from these two genes with survival traits were detected in males.
These results support our hypothesis induced by the results of experimental studies that interactions between GCN2/EIF2AK4 and CHOP/DDIT3 genes may contribute to survival in humans.
SNP-specific interaction polygenic risk scores for the rs16970024 and rs697221 SNPs
It is important to note that regulation of aging, health, and lifespan-related (survival) traits in humans may include genes and connections which are not detected in experimental studies with laboratory animals. Therefore testing associations of interactions between each of two detected SNPs and all other SNPs using available human data might result in the detection of new features of genetic human mechanism of lifespan regulation. It is because each of the detected SNPs may influence lifespan through its interactions with many other SNPs.
The number of significant associations with other SNPs and the strength of such associations may differ for each SNP. Measuring the “interacting ability” of a SNP might help better understand the contribution of a given SNP (and corresponding gene) to multifactorial regulation of lifespan. The convenient measure of such ability could be SNP-specific interaction polygenic risk score (SIPRS)[47]
Such measures have been calculated for each of the two detected SNPs. For this, genome-wide association study (GWAS)-like analysis of associations of the rs16970024 and then rs697221 SNPs with all other SNPs available in the HRS data were performed. The HRS data on white males and females combined was used in the analysis. The logistic regression model with the interaction term has been used for evaluating associations of interacting pairs of SNPs with survival traits. Education, smoking, sex, first five principal components PC1-PC5, and rs16970024 (rs697221) were used as observed covariates in the regression model. Then by using summary statistics resulted from these analyses, the rs16970024 (the rs697221) composite SIPRSs indices have been constructed using the procedure described in[47].
The construction of rs16970024 (rs697221) related indices included a set of 67,741 (66,863) interacting pairs of SNPs ranked with respect to P-values of their associations with survival trait (from smallest to the largest). Both positive and negative associations with survival traits were used in the construction of SIPRS indices. Then the properties of constructed SIPRS indices were investigated. The results of these analyses are shown in Figure 1.
Figure 1. The diagrams illustrating properties of composite indices SIPRS constructed for the rs16970024 SNP (on the left) and SIPRS for rs697221 SNP on the right. The vertical axis represents proportions of phenotypic variances (R2) explained by the composite indices. Each pillar characterizes a version of the SIPRS index corresponding to the P-value threshold (shown under the pillar at the horizontal axis). Nine pillars at each diagram correspond to nine SIPRSs indices that summarize associations of different numbers of SNP pairs with survival traits. The numbers on the top of each pillar describe properties of the corresponding index: (1) the first line on the top shows the number of SNP pairs corresponding to the P-value threshold (shown below this pillar at the horizontal axis); (2) the second line from the top shows the value of R2; (3) the third line from the top represents the P-value of association of this SIPRS index with survival trait.
The diagram on the left of Figure 1 shows that the rs16970024 SIPRS index corresponding to the threshold 5E-04 (horizontal axis under the pillar) has the smallest P-value of its association with survival trait P = 5.2E-50. This index contains 220 SNP pairs and explains 13.5% of phenotypic variance of the survival trait. The second most significant rs16970024 related index shown in this diagram corresponds to the threshold 0.001 with the P-value 1.7E-46 of its association with survival trait. This index contains 415 SNP pairs and explains 15.7% of phenotypic variance of the survival trait.
The diagram on the right of this figure shows that the rs697221 SIPRS index corresponding to the threshold 0.005 (horizontal axis under the pillar) has the smallest P-value of its association with survival trait P = 3.8E-92. This index contains 1619 SNP pairs and explains 33.4% of phenotypic variance of survival traits. The second most significant rs697221 SIPRS index corresponds to the threshold 0.001 (at horizontal axis under the pillar) with the P-value 2.7E-86 of its association with survival trait. This index contains 412 SNP pairs and explains 25.6% of phenotypic variance of the survival trait.
The R package pROC was used to calculate the areas under the receiver operating characteristics curves (AUC) for selected rs16970024 (rs697221) SIPRS indices with a corresponding threshold of 0.001. The AUC characterizes the fit of the logistic regression model describing the association of the rs16970024 (rs697221) specific SIPRS constructed from 415 (412) SNP pairs (the threshold value 0.001). The area under the curve (AUC) is 0.64 with 95% of confidence interval (0.63-0.66) for rs16970024 SIPRS, and 0.71 with 95% of confidence interval (0.70-0.72) for rs697221 SIPRS, respectively.
The properties of interacting SNP pairs most significantly associated with survival trait in which the rs16970024 SNP interacts with other SNPs are presented in Supplementary Table 1. The properties of interacting SNP pairs most significantly associated with survival trait in which the rs697221 SNP interacts with other SNPs are presented in Supplementary Table 2.
DISCUSSION
The hypothesis-free GWAS of SNPxSNP interactions, including SNPs from all selected candidate genes, is possible but involves testing many SNPxSNP interactions. The too-conservative Bonferroni correction for multiple testing often results in unjustified decisions like “we consider all associations having P-value smaller than 5.0E-03, (5.0E-04, 5.0E-05) as promising” may make the interpretation of the results of such analysis difficult. At the same time, useful insights about the potential role of interplay between specific pairs of genes in the trait of interest might be obtained from experimental studies. Testing the presence of such connection in humans can be done by estimating the association of SNPxSNP interactions with the trait using SNPs taken only from given two genes. It is important to note that, even in this case, the number of testing SNP pairs can be large enough to create problems with deciding on true-positive association. It turns out that the number of testing SNP pairs can be further reduced using the fact that many SNPs from these two genes are in LD. Making such steps increases the chances of finding true-positive associations without making assumptions which compromise statistical evidence. This paper showed how such analysis could be done. Two candidate genes have been selected, which play important roles in the ISR pathway. One, the GCN2/EIF2AK4 gene, is a sensor of amino acid starvation. Other, the CHOP/DDIT3 gene is involved in the regulation of apoptosis and autophagy. The hypothesis has been tested that the interplay between these genes may contribute to variability in human lifespan. Using data on HRS study participants, it has been found that interaction between SNP rs16970024 from the GCN2/EIF2AK4 gene and SNP rs697221 from the CHOP/DDIT3 gene is significantly associated with human survival traits for females and for males and females combined.
Geroscience and the ISR pathway
The geroscience hypothesis is that slowing down the aging process will postpone the occurrence of many age-associated health disorders, which results in increased healthspan and improved survival[117]. The idea to understand and control the individual aging process motivated many researchers to study biological mechanisms of aging and search for possible interventions that could slow down this process. One class of the interventions affecting aging has been discovered in the first half of the last century by McCay et al.[118] in experiments studying the effects of dietary restriction on lifespan in rodents. Subsequent experiments showed that a CR/DR diet is able to improve health and increase lifespan in different animal models[119,120].
More recent studies identified genetic pathways that sense disturbances in nutrients supply, regulate metabolic functions in CR/DR conditions, and influence aging, health, and lifespan[121]. A part of such regulation in laboratory animals is realized through the ISR pathway, in which interplay between the GCN2/EIF2AK4 and CHOP/DDIT3 genes dealing with the response to nutritional stress may play an essential role. Nutritional stress was an important part of life in human ancestors. This explains a major role of the sensor of the amino acid starvation (the GCN2/EIF2AK4 gene) in the ISR regulation of the processes affecting aging, health, and lifespan-related traits in response to changes in nutritional status. Experimental studies demonstrated a high potential of genes from the ISR pathway as targets for pharmacological intervention[122-128]. These studies indicated that evaluating the role of ISR pathway in human aging and lifespan may substantially improve our understanding of the factors and mechanisms initiating the development of major human age-associated health disorders.
Amino acid starvation
It was found that several mechanisms involved in sensing and regulation of response to amino acid (AA) deprivation can improve health and increase lifespan[129]. The GCN2/EIF2AK4 and mTORC1 genes are both involved in such regulation. The key role of mTOR complex 1 (mTORC1) signaling pathway regulation of aging and lifespan has been established and widely discussed in the literature[23,26,130-133]. Decreased activation of mTORC1 leads to lifespan extension in yeast, worms, flies, and mice[134].
It turns out that the mechanism is driven by the AA deficiency sensor GCN2/EIF2AK4 also influences aging, health, and lifespan/survival traits [25,135-137]. The activation of the ISR in response to nutrient starvation engages adaptive changes mediated by the induction of genes necessary to produce all the amino acids[138]. Amino acids are needed to maintain various cellular functions, including the Krebs cycle activity for ATP generation. They also provide necessary components for maintaining redox homeostasis[138]. These properties of AA regulation can be used to deal with the consequences of metabolic stress.
Insights from experimental studies should be tested using human data
The majority of information about how the interplay of genes from the ISR pathway may influence aging, health, and lifespan/survival traits is obtained in experimental studies of these traits. Even though many genetic stress response pathways are evolutionary conserved, the biological processes that involve groups of such genes in humans may differ from those developing in laboratory animals or cellular cultures. This is because, in different species, such pathways may experience species-specific modifications, acquire some, and lose other functions when adjusting to a specific biological background, nutritional differences, and external conditions. Therefore, the fact that an interplay between the GCN2/EIF2AK4 and CHOP/DDI3T genes (the members of the ISR pathway) influences lifespan/survival of laboratory animals does not mean that the same connection holds in humans. Genetic epidemiological analysis of available data on genotyped human individuals has been performed to test whether interplay between these genes is associated with human lifespan/survival. It has been found that one interacting SNP pairs taken from the GCN2/EIF2AK4 and CHOP/DDI3T genes showed a statistically significant association with human survival trait in the analyses of the HRS population of males and females combined. Sex-specific analysis revealed that a statistically significant association of interacting SNPs with survival is confirmed only in females. This result may indicate that survival in males and females are regulated using different biological mechanisms. It can also result from the fact that the population of males used in the analysis was smaller than females. This finding is an important step in the process of translation of the results of experimental studies to human applications.
Detected association of the interaction between two genes with the human survival motivates search for biological mediators of such connection
The association of the interaction between GCN2/EIF2AK4 and CHOP/DDIT3 with the human survival trait detected in statistical analysis of data does not necessarily mean that this trait is affected by the result of biochemical interaction between corresponding genetic products. Statistical analysis may capture genetic connections between two genes that could be mediated by a chain of biochemical reactions that involve products of many other genes from ISR and other signaling and metabolic pathways involved in regulating a given survival trait in humans. Identifying such mediators and evaluating their roles in the regulation of human aging, health, and survival traits could shed light on the mechanism of multifactorial regulation of these traits in humans.
The ISR pathway that activates the GCN2/EIF2AK4 and CHOP/DDIT3 along with other genes may influence the development of age-associated diseases, including cancer[59,139,140], neurodegeneration[47], diabetes[141], other[25], and through them, lifespan and survival traits. Experimental data also show that the GCN2/EIF2AK4 and CHOP/DDIT3 genes can be involved in the regulation of autophagy and apoptosis[142-146], which play a fundamental role in cancer[147,148], bacterial infections[149], other health disorders[150-156], aging[157-159]. The trade-off regulation between autophagy and apoptosis at the cellular level might be responsible for variability in lifespan[143,160]. Genes involved in such regulation are potential mediators of statistically detected association of interaction between the GCN2/EIF2AK4 and CHOP/DDIT3 genes and the survival trait.
ISR initiation may improve or deteriorate health and survival outcomes
Experimental studies provide evidence about both the positive and negative influence of the ISR initiation on health and survival traits[57,58,62]. The improvement in survival is likely to be related to the reduction of the metabolic rate in the cells at the time of cellular stress response, which is in concert with the Max Rubner’s “rate of living theory of aging”[161]. Experiments confirming the positive effects of CR/DR on aging, health, and lifespan illustrate this property[162-169]. The mechanism responsible for the positive effect on survival might be related to the fact that both genes analyzed in this paper are involved in the regulation of autophagy[142,152]. The negative correlation between basal (resting) metabolic rate and human lifespan was also detected[170,171]. All these effects were likely to be manifested, because in most of cells exposed to stress the ISR ended up by restoration of normal cellular functioning. The deleterious effects of the cellular stress response on these traits are likely to be manifested when ISR contributes to the survival of malignant cells or produces destruction of post-mitotic cells by apoptosis.
Earlier studies of the association of genetic interaction with human survival and longevity
The association of GxG interaction with human longevity has been investigated in several earlier studies. Tan et al.[172] suggested a centenarian-only approach for assessing such connections. Using data from Italian centenarians, the authors detected an association of interaction between the REN gene and the mitochondrial H haplotype with longevity. The case-control study of Han Chinese centenarians found that the interactions of SNPs from the FOXO1A and FOXO3A genes are associated with survival. This study also found that the interaction of FOXO1A and regular exercise is associated with survival traits[173]. The role of interaction between SNPs from the FOXO1A and FOXO3A genes with longevity has also been studied by implementing a novel permutation test to the data from the Danish 1905 birth cohort[174]. The analysis confirmed the association of interaction of SNPs from these genes with longevity; however, interacting SNPs detected in this study differed from those found in the study of Chinese centenarians[173]. Dato et al.[175] analyzed associations of interacting SNPs from candidate genes on longevity using data from the Danish 1905 cohort. Curk et al.[176] used an information-theoretic approach implemented in the SNPsyn software and the MDR method to select synergistic pairs of SNPs. The best combinations detected in both approaches included SNPs from IGFR and PTPN1, TP53, and ERCC2, TXNRD1 and TP53. The authors also found interacting partners: PAPPA, PTPN1, MRE11A, and PARK7 for the GHSR gene previously identified in a single-SNP association study. Ukraintseva et al. (forthcoming) investigated associations of interactions of SNPs from the group of candidate genes from aging-related pathways (IGF/FOXO growth signaling, P53/P16 apoptosis/senescence, and MTOR/SK6 mediated autophagy) with survival traits. For this, the INTERSNIP software for epistasis analysis was used in the analysis of data from the Atherosclerosis Risk in Communities study. The results of this analysis suggest that the interactions between SNPs from the IGF1R and TGFBR2 genes, as well as SNPs from the BLC2 gene, may influence human lifespan. These results were validated using data from the Cardiovascular Health Study. The results of these analyses showed that interactions between SNPs in genes from aging pathways have higher associations with survival traits than individual SNPs for the same genes[177].
Why our analysis has higher chances of rejecting the null hypothesis about the absence of true-positive associations of SNPxSNP interaction with survival trait
The hypothesis-free genome-wide association study (GWAS) of SNPxSNP interactions, including SNPs from all selected candidate genes from the ISR pathway, is possible but involves testing of many SNPxSNP interactions. The too-small P-value threshold for making the decision about the presence of true-positive association resulting from the Bonferroni correction for multiple testing often becomes responsible for statistically unjustified decisions like “we consider all associations having P-value smaller than 5.0E-03, (5.0E-04, or 5.0E-05) as promising” may make the interpretation of the results of such analysis doubtful. At the same time, useful insights about the potential role of interplay between specific pairs of genes in the trait of interest might be obtained from the results of experimental studies. Testing the presence of such connection in humans can be done by estimating the association of SNPxSNP interactions with the trait using SNPs taken only from selected two genes. It is important to note that even in this case, the number of testing SNP pairs can be large enough to create problems with deciding on true-positive association. It turns out that the number of testing SNP pairs can be further reduced using the fact that many SNPs from these two genes are in LD. Making such steps increases the chances of rejecting the null hypothesis about the absence of associations without making assumptions that compromise statistical evidence.
The use of LD for the reduction of the number of comparing SNP pairs
This paper showed how such analysis could be done. Two candidate genes have been selected, which play important roles in the ISR pathway. One, the GCN2/EIF2AK4 gene, is a sensor of amino acid starvation. Other, the CHOP/DDIT3 gene is involved in the regulation of apoptosis and autophagy. All SNPs in each gene have been divided into subsets of SNPs that are in LD with each other and whose LD measure R2 was larger or equal to a given threshold (in our case, R2 ≥ 0.1). This procedure divided all available SNP pairs into corresponding subsets. Note that the smaller is the LD threshold, the larger number of correlated SNPs that could be included in the subsets, and the smaller number of such subsets will be constructed. One representative SNP pair was selected for each subset of SNPs in LD to reduce the number of comparing SNP pairs. In our analysis, the SNP pair representatives were those who have the smallest value of the type I error among SNP pairs in this subset. This procedure resulted in 8 independent SNP pairs constructed from 8 independent SNPs from the GCN2/EIF2AK4 gene and 1 independent SNP from the CHOP/DDIT3 gene. Then this set of 8 SNP pairs was used for testing the null-hypothesis about the absence of association of the interacting SNP pairs with survival trait. Using data on HRS study participants on males and females combined, and then separately for males and females, it has been found that the null-hypothesis about the absence of association of the interaction between SNP rs16970024 from the GCN2/EIF2AK4 gene and SNP rs697221 from the CHOP/DDIT3 gene with survival trait can be rejected for females and for males and females combined, which means that the interaction of these SNPs is significantly associated with human survival trait.
The SNP specific interaction polygenic risk scores
The mechanism of multifactorial regulation of human lifespan is largely unknown. Therefore, each SNP from detected SNP pair may interact with SNPs from many other genes outside ISR pathway, and these interactions may also contribute to lifespan and survival traits. GWAS-like procedure has been used in which one of the detected SNPs is fixed, and others include all available SNPs that passed the quality control procedure. The results of this analysis were used for constructing a SNP-specific measure of collective association of many interacting SNP pairs, called the SIPRS, with survival trait. Using such an index, one gets an opportunity to evaluate collective associations with the trait of interest of different numbers of interacting SNP pairs in which a given SNP interacts with other SNPs. The approach allows for the construction of many ordered indices for a given SNP that consist of different numbers of interacting SNP pairs [Figure 1].
The properties of these indices can be compared, and the most appropriate can be selected for further analysis. It has been found that the rs16970024 SIPRS index constructed from 220 most significant interacting SNP pairs has the most significant association with survival trait with P = 5.2E-50. Index constructed from 57 most significant interacting SNP pairs is less significant with P = 2.7E-20 but contains a smaller number of SNP pairs and, hence, might be more convenient for starting a further analysis.
It has also been found that the rs697221 SIPRS constructed from 1619 most significant SNP pairs has the most significant association with survival trait with P = 3.8E-92. Index constructed from 55 SNP pairs is less significant with P = 2.E-26 [Figure 1] but contains a smaller number of SNP pairs and, hence, might be more convenient for further analysis.
Supplementary Table 1 describes properties of interacting SNP pairs in which interactions of the rs16970024 SNP with other SNPs were associated with survival trait with P-value not exceeding 9.69E-05. These interactions were used in the construction of the SIPRS for the rs16970024 SNP. Supplementary Table 2 describes properties of interacting SNP pairs in which interactions of the rs697221 SNP with other SNPs were associated with survival trait with P-value not exceeding 9.97E-05. These interactions were used in the construction of the SIPRS for the rs697221 SNP.
LIMITATIONS
Multifactorial regulation of complex traits involves interplay among many genes
Results of this study suggest that the interplay between the two key genes from the ISR pathway involved in aging, health, and lifespan traits in laboratory animals can also be involved in the regulation of human survival. This observation does not explain the entire mechanism of the multifactorial regulation of human lifespan, which can involve many other interacting genes. However, the use of only two genes of the many, allowed us to reduce the number of statistical tests and provide a proof of concept that the connection between genes and lifespan discovered in laboratory animals can also take place in humans, in the form of genetic interactions. Experimental aging studies found many other genes whose interplay may also contribute to aging, health, and survival traits. However, detecting the effects of multiple interacting genes on lifespan remains a challenging problem. Its solution requires very large sample sizes of study participants and/or advanced methods of analysis. The latter can be partly addressed by constructing and investigating the SIPRSs[47].
Dynamic regulation of aging, health, and survival traits
Individual lifespan and health status are outcomes of dynamic processes in which the interacting genetic and non-genetic factors, the strength of their interactions, and other influential variables can change with age, time, and other conditions. Genetic interaction analyses that have been performed so far, including in this study, are initial steps in addressing the multifactorial nature of complex traits and do not yet include the dynamic properties of these traits in the analysis. More efficient methods of a comprehensive statistical analysis of the dynamic polygenic regulation of the aging, health, and survival traits are urgently needed to improve our understanding of these traits substantially. Merging biodemographic methods of statistical modeling with genetic epidemiology and systems biology of aging may be a promising way to address this problem. These methods can be developed within the stress-related conceptual framework linking components of individual resistance to stresses, including robustness (vulnerability) and resilience (ability to recover)[178-182], allostatic adaptation and allostatic load[183,184] with health and survival outcomes.
From association to causality
The associations of interacting SNP pairs with survival traits evaluated in this paper do not necessarily describe causal connections. This limitation is common for statistical methods used in genetic analyses of observational data. Information about causal connections is needed for developing intervention therapies, testing candidate medications. Testing causality of connections detected in the analysis of observational data can be performed using methods of mediation analysis[185] and Mendelian randomization[186]. These methods have their own limitations[187].
Lack of information about cellular stressors in human data
The use of the stress-related conceptual framework in studies of multifactorial mechanisms of lifespan regulation would benefit from information about biomarkers characterizing cellular stressors, as well as about factors and conditions capable of affecting variability (e.g., strength, duration) of the cellular stress response. Since many human data on aging, health, and lifespan related traits have limited information about such stressors, factors, and conditions, some useful insights and ideas about possible stressors can be obtained indirectly from the estimates of the roles of specific genetic sensors of cellular stress signals in lifespan regulation obtained in data analyses. This is because each such sensor recognizes and responds to specific (sometimes overlapping) groups of stress signals.
Genetic interactions: statistical vs. biological epistasis
Genetic interactions may produce paradoxical results in the genetic association analyses of complex traits[107,188-191]. One should, however, distinguish between biological and statistical epistasis. In biological experiments, epistasis may be detected as a result of proteins’ interactions, in which the effect of one protein can mask the effect of another protein on the phenotype of interest[107]. In genetic epidemiology, the effects of genetic interactions on the trait of interest are evaluated using regression models with the interaction terms[46]. Since these genetic epidemiology methods differ from those used in detecting the effects of epistasis in biological experiments, the results of such analyses may also have different interpretations[107,192]. Associations of genetic interactions with survival traits detected in epidemiological studies may reflect synergistic or antagonistic effects of genes whose products do not directly influence each other and, therefore, will not necessarily be detected in studies of biological epistasis. In contrast to biological epistasis, statistical interactions can capture connections between genes mediated by the chain of intermediate genetic products. Identification of such intermediate genes and evaluation of their roles in aging, health, and lifespan may require new experimental studies. The detected statistical association does not exclude the presence of the effect of biological epistasis between two genetic products on human lifespan. However, the association of statistical genetic interaction with survival may be detected even in the absence of direct biological interaction between genetic products. This might be the case when such interaction and connection with the trait are mediated by other genes.
Long road to understanding multifactorial mechanism regulating human lifespan
The results of this paper do not provide us with all the details about mechanisms of multifactorial regulation of human lifespan. They, rather, describe a way to become more informative about it. An important question that was not addressed in this study is how the confirmed (or newly detected) information about multifactorial regulation of aging, health, and survival in humans should be integrated to explain health and survival outcomes? To be seful in practice, mechanisms of such integration should have quantitative description, e.g., in the form of a computer model capable of describing the response of the body (cells, organs, systems) to specific challenges. A promising tool for addressing this problem could be further development of dynamic stochastic modeling of human mortality and aging, which successfully used in the analysis of longitudinal data[182].
Conclusion
Experimental studies pointed to a fundamental role of the interplay between the GCN2/EIF2AK4 (involved in sensing cellular stress signals) and CHOP/DDIT3 (involved in apoptosis and autophagy), as well as other genes from the ISR pathway, in aging, health, and lifespan of laboratory animals[25,193], which encourages clarifying the role of interactions among these genes in respective human traits.
Results of this study support our hypothesis that the interplay between GCN2/EIF2AK4 and CHOP/DDIT3 genes involved in the ISR pathway may influence lifespan in humans. This is a “proof of concept” research and a step forward to translating the evidence about mechanisms of lifespan regulation found in laboratory animals to humans.
Individual differences in the exposure to stressful conditions, in the access to health care, the polymorphisms in genes that sense cellular stress signals, and in other genes involved in cellular stress response, together with aging-related changes in robustness and resilience, are likely to be important sources of disparity and heterogeneity in health, lifespan and survival outcomes. Being evaluated, these characteristics can be used in predicting the future burden of aging-related diseases under different scenarios/strategies of reducing disparities, improving access to health care facilities and medical advances for the groups of individuals having different genetic backgrounds and exposed to distinct environmental and living conditions.
DECLARATIONS
Authors’ contributionsDeveloped conceptual idea of this study: Yashin AI, Ukraintseva S
Prepared the first draft of the paper: Yashin AI
Developed computational structure and performed data analyses: Wu D
Provided useful comments and interpretation of the research results: Ukraintseva S, Arbeev K,
Prepared the data for analysis: Duan M, Bagley O
Read the paper, provided valuable suggestions and comments, which contributed to the ultimate version of the manuscript: Yashin AI, Wu DQ, Arbeev K, Yashkin AP, Akushevich I, Bagley O, Duan M,
Not applicable.
Financial support and sponsorshipThis study was supported by the NIH/NIA grants RF1AG046860, R01AG062623, and R01AG066133. The Health and Retirement Study (HRS) data is sponsored by the National Institute on Aging (grant number U01AG009740) and is conducted by the University of Michigan. The HRS genetic data is sponsored by the National Institute on Aging (grants U01AG009740, RC2AG036495, and RC4AG039029) and was conducted by the University of Michigan. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
2. Johnson TE, Wood WB. Genetic analysis of life-span in Caenorhabditis elegans. Proc Natl Acad Sci U S A 1982;79:6603-7.
3. Johnson TE, Conley WL, Keller ML. Long-lived lines of Caenorhabditis elegans can be used to establish predictive biomarkers of aging. Exp Gerontol 1988;23:281-95.
4. Friedman DB, Johnson TE. Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J Gerontol 1988;43:B102-9.
6. Partridge L, Pletcher SD, Mair W. Dietary restriction, mortality trajectories, risk and damage. Mech Ageing Dev 2005;126:35-41.
7. Masoro EJ. Caloric restriction and aging: controversial issues. J Gerontol A Biol Sci Med Sci 2006;61:14-9.
8. Archer CR, Royle N, South S, Selman C, Hunt J. Nutritional geometry provides food for thought. J Gerontol A Biol Sci Med Sci 2009;64:956-9.
10. Couteur DG, Solon-Biet S, Cogger VC, et al. The impact of low-protein high-carbohydrate diets on aging and lifespan. Cell Mol Life Sci 2016;73:1237-52.
11. Seidelmann SB, Claggett B, Cheng S, et al. Dietary carbohydrate intake and mortality: a prospective cohort study and meta-analysis. Lancet Public Health 2018;3:e419-e28.
12. Johnson AA, Stolzing A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 2019;18:e13048.
13. Yoshida S, Yamahara K, Kume S, et al. Role of dietary amino acid balance in diet restriction-mediated lifespan extension, renoprotection, and muscle weakness in aged mice. Aging Cell 2018;17:e12796.
14. Kitada M, Ogura Y, Monno I, Koya D. The impact of dietary protein intake on longevity and metabolic health. EBioMedicine 2019;43:632-40.
15. Lee D, Hwang W, Artan M, Jeong DE, Lee SJ. Effects of nutritional components on aging. Aging Cell 2015;14:8-16.
16. Arganda S, Bouchebti S, Bazazi S, et al. Parsing the life-shortening effects of dietary protein: effects of individual amino acids. Proc Biol Sci 2017:284.
17. Ames BN. Prolonging healthy aging: longevity vitamins and proteins. Proc Natl Acad Sci U S A 2018;115:10836-44.
18. Altamura S, Muckenthaler MU. Iron toxicity in diseases of aging: Alzheimer's disease, Parkinson's disease and atherosclerosis. J Alzheimers Dis 2009;16:879-95.
19. Nie Y, Luo F. Dietary fiber: an opportunity for a global control of hyperlipidemia. Oxid Med Cell Longev 2021;2021:5542342.
20. Klass MR. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Ageing Dev 1983;22:279-86.
21. Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 2010;9:92-5.
22. Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 2013;14:133-9.
23. Goberdhan DC, Wilson C, Harris AL. Amino acid sensing by mTORC1: intracellular transporters mark the spot. Cell Metab 2016;23:580-9.
24. Averous J, Lambert-Langlais S, Mesclon F, et al. GCN2 contributes to mTORC1 inhibition by leucine deprivation through an ATF4 independent mechanism. Sci Rep 2016;6:27698.
25. Falcón P, Escandón M, Brito Á, Matus S. Nutrient sensing and redox balance: GCN2 as a new integrator in aging. Oxid Med Cell Longev 2019;2019:5730532.
26. Papadopoli D, Boulay K, Kazak L, et al. mTOR as a central regulator of lifespan and aging. F1000Res 2019:8.
27. Uppu S, Krishna A, Gopalan RP. A review on methods for detecting SNP interactions in high-dimensional genomic data. IEEE/ACM Trans Comput Biol Bioinform 2018;15:599-612.
28. Liu L, Wong KC. Epistasis analysis: classification through machine learning methods. Methods Mol Biol 2021;2212:337-45.
29. Manavalan R, Priya S. Genetic interactions effects for cancer disease identification using computational models: a review. Med Biol Eng Comput 2021;59:733-58.
30. Slim L, Chatelain C, Azencott CA, Vert JP. Novel methods for epistasis detection in genome-wide association studies. PLoS One 2020;15:e0242927.
31. Pan Q, Hu T, Moore JH. Epistasis, complexity, and multifactor dimensionality reduction. Methods Mol Biol 2013;1019:465-77.
32. Abegaz F, Van Lishout F, Mahachie John JM, et al. Performance of model-based multifactor dimensionality reduction methods for epistasis detection by controlling population structure. BioData Min 2021;14:16.
33. Yang CH, Lin YD, Chuang LY. Class balanced multifactor dimensionality reduction to detect gene-gene interactions. IEEE/ACM Trans Comput Biol Bioinform 2020;17:71-81.
34. Chen G, Yuan A, Cai T, et al. Measuring gene-gene interaction using Kullback-Leibler divergence. Ann Hum Genet 2019;83:405-17.
35. Malten J, König IR. Modified entropy-based procedure detects gene-gene-interactions in unconventional genetic models. BMC Med Genomics 2020;13:65.
36. Borzou A, Sadygov RG. A novel estimator of the interaction matrix in Graphical Gaussian Model of omics data using the entropy of non-equilibrium systems. Bioinformatics 2021;37:837-44.
37. Chicco D, Faultless T. Brief survey on machine learning in epistasis. Methods Mol Biol 2021;2212:169-79.
38. Petinrin OO, Wong KC. Protocol for epistasis detection with machine learning using genepi package. Methods Mol Biol 2021;2212:291-305.
39. Lin HY, Chen DT, Huang PY, et al. SNP interaction pattern identifier (SIPI): an intensive search for SNP-SNP interaction patterns. Bioinformatics 2017;33:822-33.
40. Sun R, Xia X, Chong KC, Zee BC, Wu WKK, Wang MH. wtest: an integrated R package for genetic epistasis testing. BMC Med Genomics 2019;12:180.
41. Cao X, Liu J, Guo M, Wang J. HiSSI: high-order SNP-SNP interactions detection based on efficient significant pattern and differential evolution. BMC Med Genomics 2019;12:139.
42. Carmelo VAO, Kogelman LJA, Madsen MB, Kadarmideen HN. WISH-R- a fast and efficient tool for construction of epistatic networks for complex traits and diseases. BMC Bioinformatics 2018;19:277.
43. Chang YC, Wu JT, Hong MY, et al. GenEpi: gene-based epistasis discovery using machine learning. BMC Bioinformatics 2020;21:68.
44. Huang Y, Wuchty S, Przytycka TM. eQTL Epistasis - challenges and computational approaches. Front Genet 2013;4:51.
45. Kwon M, Leem S, Yoon J, Park T. GxGrare: gene-gene interaction analysis method for rare variants from high-throughput sequencing data. BMC Syst Biol 2018;12:19.
46. Herold C, Steffens M, Brockschmidt FF, Baur MP, Becker T. INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics 2009;25:3275-81.
47. Yashin AI, Wu D, Arbeev K, et al. Interplay between stress-related genes may influence Alzheimer's disease development: the results of genetic interaction analyses of human data. Mech Ageing Dev 2021;196:111477.
48. Niel C, Sinoquet C, Dina C, Rocheleau G. A survey about methods dedicated to epistasis detection. Front Genet 2015;6:285.
49. Costa-Mattioli M, Walter P. The integrated stress response: from mechanism to disease. Science 2020:368.
50. Bruhat A, Jousse C, Wang XZ, Ron D, Ferrara M, Fafournoux P. Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both transcriptional and post-transcriptional levels. J Biol Chem 1997;272:17588-93.
51. Masuda M, Miyazaki-Anzai S, Levi M, Ting TC, Miyazaki M. PERK-eIF2α-ATF4-CHOP signaling contributes to TNFα-induced vascular calcification. J Am Heart Assoc 2013;2:e000238.
52. Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016;17:1374-95.
53. Wang L, Tang W, Jiang T, et al. Endoplasmic reticulum stress is involved in the neuroprotective effect of propofol. Neurochem Res 2014;39:1741-52.
54. Oliveira MM, Lourenco MV. Integrated stress response: connecting ApoE4 to memory impairment in Alzheimer's disease. J Neurosci 2016;36:1053-5.
55. Bond S, Lopez-Lloreda C, Gannon PJ, Akay-Espinoza C, Jordan-Sciutto KL. The integrated stress response and phosphorylated eukaryotic initiation factor 2α in neurodegeneration. J Neuropathol Exp Neurol 2020;79:123-43.
56. Girardin SE, Cuziol C, Philpott DJ, Arnoult D. The eIF2α kinase HRI in innate immunity, proteostasis, and mitochondrial stress. Febs j :202010.1111/febs.15553.
57. Postnikoff SDL, Johnson JE, Tyler JK. The integrated stress response in budding yeast lifespan extension. Microb Cell 2017;4:368-75.
58. Derisbourg MJ, Wester LE, Baddi R, Denzel MS. Mutagenesis screen uncovers lifespan extension through integrated stress response inhibition without reduced mRNA translation. Nat Commun 2021;12:1678.
59. Wang Y, Ning Y, Alam GN, et al. Amino acid deprivation promotes tumor angiogenesis through the GCN2/ATF4 pathway. Neoplasia 2013;15:989-97.
60. Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The role of the PERK/eIF2α/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr Mol Med 2016;16:533-44.
61. Ye J, Kumanova M, Hart LS, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. Embo j 2010;29:2082-96.
62. Emanuelli G, Nassehzadeh-Tabriz N, Morrell NW, Marciniak SJ. The integrated stress response in pulmonary disease. Eur Respir Rev 2020:29.
63. Fuster JJ. Integrated stress response inhibition in atherosclerosis: preventing the stressed-out plaque. J Am Coll Cardiol 2019;73:1170-2.
64. Onat UI, Yildirim AD, Tufanli Ö, et al. Intercepting the lipid-induced integrated stress response reduces atherosclerosis. J Am Coll Cardiol 2019;73:1149-69.
65. Xu X, Krumm C, So JS, et al. Preemptive activation of the integrated stress response protects mice from diet-induced obesity and insulin resistance by fibroblast growth factor 21 induction. Hepatology 2018;68:2167-81.
66. Henkel AS. Harnessing the integrated stress response to counteract metabolic disease. Hepatology 2018;68:2056-8.
67. Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: their structures and functions. Cell Mol Life Sci 2013;70:3493-511.
68. Gáll T, Balla G, Balla J. Heme, heme oxygenase, and endoplasmic reticulum stress-a new insight into the pathophysiology of vascular diseases. Int J Mol Sci 2019:20.
69. Rodrigues L, Graça RSF, Carneiro LAM. Integrated stress responses to bacterial pathogenesis patterns. Front Immunol 2018;9:1306.
70. Saelens X, Kalai M, Vandenabeele P. Translation inhibition in apoptosis: caspase-dependent PKR activation and eIF2-alpha phosphorylation. J Biol Chem 2001;276:41620-8.
71. Taylor SS, Haste NM, Ghosh G. PKR and eIF2alpha: integration of kinase dimerization, activation, and substrate docking. Cell 2005;122:823-5.
72. Chesnokova E, Bal N, Kolosov P. Kinases of eIF2a switch translation of mRNA subset during neuronal plasticity. Int J Mol Sci 2017:18.
73. Gal-Ben-Ari S, Barrera I, Ehrlich M, Rosenblum K. PKR: a kinase to remember. Front Mol Neurosci 2018;11:480.
74. Elvira R, Cha SJ, Noh GM, Kim K, Han J. PERK-mediated eIF2α phosphorylation contributes to the protection of dopaminergic neurons from chronic heat stress in drosophila. Int J Mol Sci 2020:21.
75. Klaus S, Ost M. Mitochondrial uncoupling and longevity - a role for mitokines? Exp Gerontol 2020;130:110796.
76. Wang R, McGrath BC, Kopp RF, et al. Insulin secretion and Ca2+ dynamics in β-cells are regulated by PERK (EIF2AK3) in concert with calcineurin. J Biol Chem 2013;288:33824-36.
77. Wang R, Munoz EE, Zhu S, McGrath BC, Cavener DR. Perk gene dosage regulates glucose homeostasis by modulating pancreatic β-cell functions. PLoS One 2014;9:e99684.
78. Kim MJ, Min SH, Shin SY, et al. Attenuation of PERK enhances glucose-stimulated insulin secretion in islets. J Endocrinol 2018;236:125-36.
79. Balsa E, Soustek MS, Thomas A, et al. ER and nutrient stress promote assembly of respiratory chain supercomplexes through the PERK-eIF2α axis. Mol Cell 2019;74:877-90.e6.
80. Endres K, Reinhardt S. ER-stress in Alzheimer's disease: turning the scale? Am J Neurodegener Dis 2013;2:247-65.
81. Wong TH, van der Lee SJ, van Rooij JGJ, et al. EIF2AK3 variants in Dutch patients with Alzheimer's disease. Neurobiol Aging 2019;73:229.e11-e18.
82. Hughes D, Mallucci GR. The unfolded protein response in neurodegenerative disorders - therapeutic modulation of the PERK pathway. FEBS J 2019;286:342-55.
83. Rozpędek-Kamińska W, Siwecka N, Wawrzynkiewicz A, et al. The PERK-dependent molecular mechanisms as a novel therapeutic target for neurodegenerative diseases. Int J Mol Sci 2020:21.
84. Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell 2005;16:5493-501.
86. Chikashige Y, Kato H, Thornton M, et al. Gcn2 eIF2α kinase mediates combinatorial translational regulation through nucleotide motifs and uORFs in target mRNAs. Nucleic Acids Res 2020;48:8977-92.
87. Yuan F, Jiang H, Yin H, et al. Activation of GCN2/ATF4 signals in amygdalar PKC-δ neurons promotes WAT browning under leucine deprivation. Nat Commun 2020;11:2847.
88. Augusto L, Amin PH, Wek RC, Sullivan WJ, Jr. Regulation of arginine transport by GCN2 eIF2 kinase is important for replication of the intracellular parasite Toxoplasma gondii. PLoS Pathog 2019;15:e1007746.
89. Hayner JN, Shan J, Kilberg MS. Regulation of the ATF3 gene by a single promoter in response to amino acid availability and endoplasmic reticulum stress in human primary hepatocytes and hepatoma cells. Biochim Biophys Acta Gene Regul Mech 2018;1861:72-9.
90. Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000;6:1099-108.
91. Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab 2009;20:436-43.
92. Rasmussen BB, Adams CM. ATF4 is a fundamental regulator of nutrient sensing and protein turnover. J Nutr 2020;150:979-80.
93. Tian F, Zhao J, Bu S, et al. KLF6 induces apoptosis in human lens epithelial cells through the ATF4-ATF3-CHOP axis. Drug Des Devel Ther 2020;14:1041-55.
94. Choy MS, Yusoff P, Lee IC, et al. Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep 2015;11:1885-91.
95. Joo JH, Ueda E, Bortner CD, Yang XP, Liao G, Jetten AM. Farnesol activates the intrinsic pathway of apoptosis and the ATF4-ATF3-CHOP cascade of ER stress in human T lymphoblastic leukemia Molt4 cells. Biochem Pharmacol 2015;97:256-68.
97. Pike LR, Phadwal K, Simon AK, Harris AL. ATF4 orchestrates a program of BH3-only protein expression in severe hypoxia. Mol Biol Rep 2012;39:10811-22.
98. Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem 2010;285:33165-74.
99. Rzymski T, Milani M, Pike L, et al. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 2010;29:4424-35.
100. Frank CL, Ge X, Xie Z, Zhou Y, Tsai LH. Control of activating transcription factor 4 (ATF4) persistence by multisite phosphorylation impacts cell cycle progression and neurogenesis. J Biol Chem 2010;285:33324-37.
101. Mukherjee D, Bercz LS, Torok MA, Mace TA. Regulation of cellular immunity by activating transcription factor 4. Immunol Lett 2020;228:24-34.
102. Narita T, Ri M, Masaki A, et al. Lower expression of activating transcription factors 3 and 4 correlates with shorter progression-free survival in multiple myeloma patients receiving bortezomib plus dexamethasone therapy. Blood Cancer J 2015;5:e373.
103. Jousse C, Deval C, Maurin AC, et al. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J Biol Chem 2007;282:15851-61.
104. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nature Protocols 2010;5:1564-73.
105. Marees AT, de Kluiver H, Stringer S, et al. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res 2018;27:e1608.
106. Akushevich I, Kravchenko J, Ukraintseva S, Arbeev K, Yashin AI. Age patterns of incidence of geriatric disease in the U.S. elderly population: medicare-based analysis. J Am Geriatr Soc 2012;60:323-7.
107. Ukraintseva S, Yashin A, Arbeev K, et al. Puzzling role of genetic risk factors in human longevity: "risk alleles" as pro-longevity variants. Biogerontology 2016;17:109-27.
108. Chasioti D, Yan J, Nho K, Saykin AJ. Progress in polygenic composite scores in Alzheimer's and other complex diseases. Trends Genet 2019;35:371-82.
109. Cruchaga C, Del-Aguila JL, Saef B, et al. Polygenic risk score of sporadic late-onset Alzheimer's disease reveals a shared architecture with the familial and early-onset forms. Alzheimers Dement 2018;14:205-14.
110. Leonenko G, Sims R, Shoai M, et al. Polygenic risk and hazard scores for Alzheimer's disease prediction. Ann Clin Transl Neurol 2019;6:456-65.
111. Tesi N, van der Lee SJ, Hulsman M, et al. Polygenic risk score of longevity predicts longer survival across an age continuum. J Gerontol A Biol Sci Med Sci 2021;76:750-9.
112. Zeng Y, Nie C, Min J, et al. Sex differences in genetic associations with longevity. JAMA Netw Open 2018;1:e181670.
113. Yashin AI, Wu D, Arbeev KG, Ukraintseva SV. Joint influence of small-effect genetic variants on human longevity. Aging (Albany NY) 2010;2:612-20.
114. Yashin AI, Wu D, Arbeev KG, Stallard E, Land KC, Ukraintseva SV. How genes influence life span: the biodemography of human survival. Rejuvenation Res 2012;15:374-80.
115. Yashin AI, Wu D, Arbeev KG, Ukraintseva SV. Polygenic effects of common single-nucleotide polymorphisms on life span: when association meets causality. Rejuvenation Res 2012;15:381-94.
116. Ala-Korpela M, Holmes MV. Polygenic risk scores and the prediction of common diseases. Int J Epidemiol 2020;49:1-3.
117. Yabluchanskiy A, Ungvari Z, Csiszar A, Tarantini S. Advances and challenges in geroscience research: an update. Physiol Int 2018;105:298-308.
118. McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition 1989;5:155-71; discussion 72.
119. Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell 2015;161:106-18.
120. Balasubramanian P, Mattison JA, Anderson RM. Nutrition, metabolism, and targeting aging in nonhuman primates. Ageing Res Rev 2017;39:29-35.
121. Cummings NE, Lamming DW. Regulation of metabolic health and aging by nutrient-sensitive signaling pathways. Mol Cell Endocrinol 2017;455:13-22.
122. Edick AM, Audette J, Burgos SA. CRISPR-Cas9-mediated knockout of GCN2 reveals a critical role in sensing amino acid deprivation in bovine mammary epithelial cells. J Dairy Sci 2021;104:1123-35.
123. Xia X, Lei L, Qin W, Wang L, Zhang G, Hu J. GCN2 controls the cellular checkpoint: potential target for regulating inflammation. Cell Death Discov 2018;4:20.
124. Hu G, Yu Y, Tang YJ, Wu C, Long F, Karner CM. The amino acid sensor Eif2ak4/GCN2 is required for proliferation of osteoblast progenitors in mice. J Bone Miner Res 2020;35:2004-14.
125. Revelo XS, Winer S, Winer DA. Starving intestinal inflammation with the amino acid sensor GCN2. Cell Metab 2016;23:763-5.
126. Anand AA, Walter P. Structural insights into ISRIB, a memory-enhancing inhibitor of the integrated stress response. FEBS J 2020;287:239-45.
127. Chu HS, Peterson C, Jun A, Foster J. Targeting the integrated stress response in ophthalmology. Curr Eye Res 2021;46:1075-88.
128. Zyryanova AF, Kashiwagi K, Rato C, et al. ISRIB blunts the integrated stress response by allosterically antagonising the inhibitory effect of phosphorylated eIF2 on eIF2B. Mol Cell 2021;81:88-103.e6.
129. Fontana L, Partridge L, Longo VD. Extending healthy life span - from yeast to humans. Science 2010;328:321-6.
130. Antikainen H, Driscoll M, Haspel G, Dobrowolski R. TOR-mediated regulation of metabolism in aging. Aging Cell 2017;16:1219-33.
132. Jung J, Genau HM, Behrends C. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol 2015;35:2479-94.
133. Kucheryavenko O, Nelson G, von Zglinicki T, Korolchuk VI, Carroll B. The mTORC1-autophagy pathway is a target for senescent cell elimination. Biogerontology 2019;20:331-5.
134. Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature 2013;493:338-45.
135. Longchamp A, Mirabella T, Arduini A, et al. Amino acid restriction triggers angiogenesis via GCN2/ATF4 regulation of VEGF and H(2)S production. Cell 2018;173:117-29.e14.
136. Pathak SS, Liu D, Li T, et al. The eIF2α Kinase GCN2 modulates period and rhythmicity of the circadian clock by translational control of Atf4. Neuron 2019;104:724-35.e6.
137. Pereira CM, Filev R, Dubiela FP, et al. The GCN2 inhibitor IMPACT contributes to diet-induced obesity and body temperature control. PLoS One 2019;14:e0217287.
138. Harding HP, Zhang Y, Zeng H, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003;11:619-33.
139. Rashidi A, Miska J, Lee-Chang C, et al. GCN2 is essential for CD8(+) T cell survival and function in murine models of malignant glioma. Cancer Immunol Immunother 2020;69:81-94.
140. Schmidt S, Gay D, Uthe FW, et al. A MYC-GCN2-eIF2α negative feedback loop limits protein synthesis to prevent MYC-dependent apoptosis in colorectal cancer. Nat Cell Biol 2019;21:1413-24.
141. Kanno A, Asahara SI, Furubayashi A, et al. GCN2 regulates pancreatic β cell mass by sensing intracellular amino acid levels. JCI Insight 2020;5:e128820.
142. B'Chir W, Chaveroux C, Carraro V, et al. Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation. Cell Signal 2014;26:1385-91.
143. B'Chir W, Maurin AC, Carraro V, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 2013;41:7683-99.
144. Carroll B, Korolchuk VI, Sarkar S. Amino acids and autophagy: cross-talk and co-operation to control cellular homeostasis. Amino Acids 2015;47:2065-88.
145. Humeau J, Bezu L, Kepp O, Kroemer G. EIF2α phosphorylation: a hallmark of both autophagy and immunogenic cell death. Mol Cell Oncol 2020;7:1776570.
146. Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010;40:280-93.
147. Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev 2016;30:1913-30.
148. Acevo-Rodríguez PS, Maldonado G, Castro-Obregón S, Hernández G. Autophagy regulation by the translation machinery and its implications in cancer. Front Oncol 2020;10:322.
149. Castrejón-Jiménez NS, Leyva-Paredes K, Hernández-González JC, Luna-Herrera J, García-Pérez BE. The role of autophagy in bacterial infections. Biosci Trends 2015;9:149-59.
150. Nakahira K, Pabon Porras MA, Choi AM. Autophagy in pulmonary diseases. Am J Respir Crit Care Med 2016;194:1196-207.
151. Andhavarapu S, Mubariz F, Arvas M, Bever C, Jr. , Makar TK. Interplay between ER stress and autophagy: a possible mechanism in multiple sclerosis pathology. Exp Mol Pathol 2019;108:183-90.
152. Bretin A, Carrière J, Dalmasso G, et al. Activation of the EIF2AK4-EIF2A/eIF2α-ATF4 pathway triggers autophagy response to Crohn disease-associated adherent-invasive Escherichia coli infection. Autophagy 2016;12:770-83.
153. Ryter SW, Lee SJ, Smith A, Choi AM. Autophagy in vascular disease. Proc Am Thorac Soc 2010;7:40-7.
155. Tan P, Ye Y, Mao J, He L. Autophagy and immune-related diseases. Adv Exp Med Biol 2019;1209:167-79.
156. Wu J, Ye J, Kong W, Zhang S, Zheng Y. Programmed cell death pathways in hearing loss: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif 2020;53:e12915.
157. Escobar KA, Cole NH, Mermier CM, VanDusseldorp TA. Autophagy and aging: maintaining the proteome through exercise and caloric restriction. Aging Cell 2019;18:e12876.
158. Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta 2009;1793:664-73.
159. Rajawat YS, Hilioti Z, Bossis I. Aging: central role for autophagy and the lysosomal degradative system. Ageing Res Rev 2009;8:199-213.
160. Hansen M, Rubinsztein DC, Walker DW. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol 2018;19:579-93.
161. Rubner M. Machinery of metabolism. JAMA 1916;66:1111.
162. Fontana L. The scientific basis of caloric restriction leading to longer life. Curr Opin Gastroenterol 2009;25:144-50.
163. Ferrucci L, Schrack JA, Knuth ND, Simonsick EM. Aging and the energetic cost of life. J Am Geriatr Soc 2012;60:1768-9.
164. Al-Regaiey KA. The effects of calorie restriction on aging: a brief review. Eur Rev Med Pharmacol Sci 2016;20:2468-73.
165. Bartke A, Evans TR, Musters CJM. Anti-aging interventions affect lifespan variability in sex, strain, diet and drug dependent fashion. Aging (Albany NY) 2019;11:4066-74.
166. Cantó C, Auwerx J. Calorie restriction: is AMPK a key sensor and effector? Physiology (Bethesda) 2011;26:214-24.
167. Colman RJ, Anderson RM. Nonhuman primate calorie restriction. Antioxid Redox Signal 2011;14:229-39.
168. Flanagan EW, Most J, Mey JT, Redman LM. Calorie restriction and aging in humans. Annu Rev Nutr 2020;40:105-33.
169. Green CL, Lamming DW. Regulation of metabolic health by essential dietary amino acids. Mech Ageing Dev 2019;177:186-200.
170. Ruggiero C, Metter EJ, Melenovsky V, et al. High basal metabolic rate is a risk factor for mortality: the Baltimore Longitudinal Study of Aging. J Gerontol A Biol Sci Med Sci 2008;63:698-706.
171. Jumpertz R, Hanson RL, Sievers ML, Bennett PH, Nelson RG, Krakoff J. Higher energy expenditure in humans predicts natural mortality. J Clin Endocrinol Metab 2011;96:E972-6.
172. Tan Q, Yashin AI, Bladbjerg EM, et al. A case-only approach for assessing gene by sex interaction in human longevity. J Gerontol A Biol Sci Med Sci 2002;57:B129-33.
173. Zeng Y, Cheng L, Chen H, et al. Effects of FOXO genotypes on longevity: a biodemographic analysis. J Gerontol A Biol Sci Med Sci 2010;65:1285-99.
174. Tan Q, Soerensen M, Kruse TA, Christensen K, Christiansen L. A novel permutation test for case-only analysis identifies epistatic effects on human longevity in the FOXO gene family. Aging Cell 2013;12:690-4.
175. Dato S, Soerensen M, De Rango F, et al. The genetic component of human longevity: new insights from the analysis of pathway-based SNP-SNP interactions. Aging Cell 2018;17:e12755.
176. Curk T, Rot G, Zupan B. SNPsyn: detection and exploration of SNP-SNP interactions. Nucleic Acids Res 2011;39:W444-9.
177. Ukraintseva S, Duan M, Arbeev K, et al. Interactions between genes from aging pathways may influence human lifespan and improve animal to human translation. Front Cell Dev Biol 2021;9:692020.
178. Ukraintseva S, Arbeev K, Duan M, et al. Decline in biological resilience as key manifestation of aging: potential mechanisms and role in health and longevity. Mech Ageing Dev 2021;194:111418.
179. Ukraintseva S, Yashin AI, Arbeev KG. Resilience versus robustness in aging. J Gerontol A Biol Sci Med Sci 2016;71:1533-4.
180. Galvin A, Ukraintseva S, Arbeev K, Feitosa M, Christensen K. Physical robustness and resilience among long-lived female siblings: a comparison with sporadic long-livers. Aging (Albany NY) 2020;12:15157-68.
181. Arbeev KG, Akushevich I, Kulminski AM, Ukraintseva SV, Yashin AI. Biodemographic analyses of longitudinal data on aging, health, and longevity: recent advances and future perspectives. Adv Geriatr 2014;2014:957073.
182. Arbeev KG, Ukraintseva SV, Bagley O, et al. "Physiological Dysregulation" as a promising measure of robustness and resilience in studies of aging and a new indicator of preclinical disease. J Gerontol A Biol Sci Med Sci 2019;74:462-8.
183. Yashin AI, Arbeev KG, Akushevich I, et al. The quadratic hazard model for analyzing longitudinal data on aging, health, and the life span. Phys Life Rev 2012;9:177-88.
184. Yashin AI, Arbeev KG, Arbeeva LS, et al. How the effects of aging and stresses of life are integrated in mortality rates: insights for genetic studies of human health and longevity. Biogerontology 2016;17:89-107.
185. Agler R, De Boeck P. On the interpretation and use of mediation: multiple perspectives on mediation analysis. Front Psychol 2017;8:1984.
186. Grover S, Del Greco MF, König IR. Evaluating the current state of Mendelian randomization studies: a protocol for a systematic review on methodological and clinical aspects using neurodegenerative disorders as outcome. Syst Rev 2018;7:145.
187. Allman PH, Aban IB, Tiwari HK, Cutter GR. An introduction to Mendelian randomization with applications in neurology. Multiple sclerosis and related disorders 2018;24:72-8.
188. Yashin AI, De Benedictis G, Vaupel JW, et al. Genes, demography, and life span: the contribution of demographic data in genetic studies on aging and longevity. Am J Hum Genet 1999;65:1178-93.
189. Yashin AI, De Benedictis G, Vaupel JW, et al. Genes and longevity: lessons from studies of centenarians. J Gerontol A Biol Sci Med Sci 2000;55:B319-28.
190. Bergman A, Atzmon G, Ye K, MacCarthy T, Barzilai N. Buffering mechanisms in aging: a systems approach toward uncovering the genetic component of aging. PLoS Comput Biol 2007;3:e170.
191. Beekman M, Nederstigt C, Suchiman HE, et al. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. Proc Natl Acad Sci U S A 2010;107:18046-9.
192. Ebbert MT, Ridge PG, Kauwe JS. Bridging the gap between statistical and biological epistasis in Alzheimer's disease. Biomed Res Int 2015;2015:870123.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Yashin AI, Wu D, Arbeev K, Yashkin AP, Akushevich I, Bagley O, Duan M, Ukraintseva S. Roles of interacting stress-related genes in lifespan regulation: insights for translating experimental findings to humans. J Transl Genet Genom 2021;5:357-79. http://dx.doi.org/10.20517/jtgg.2021.26
AMA Style
Yashin AI, Wu D, Arbeev K, Yashkin AP, Akushevich I, Bagley O, Duan M, Ukraintseva S. Roles of interacting stress-related genes in lifespan regulation: insights for translating experimental findings to humans. Journal of Translational Genetics and Genomics. 2021; 5(4): 357-79. http://dx.doi.org/10.20517/jtgg.2021.26
Chicago/Turabian Style
Yashin, Anatoliy I., Deqing Wu, Konstantin Arbeev, Arseniy P. Yashkin, Igor Akushevich, Olivia Bagley, Matt Duan, Svetlana Ukraintseva. 2021. "Roles of interacting stress-related genes in lifespan regulation: insights for translating experimental findings to humans" Journal of Translational Genetics and Genomics. 5, no.4: 357-79. http://dx.doi.org/10.20517/jtgg.2021.26
ACS Style
Yashin, AI.; Wu D.; Arbeev K.; Yashkin AP.; Akushevich I.; Bagley O.; Duan M.; Ukraintseva S. Roles of interacting stress-related genes in lifespan regulation: insights for translating experimental findings to humans. J. Transl. Genet. Genom. 2021, 5, 357-79. http://dx.doi.org/10.20517/jtgg.2021.26
About This Article
Copyright
Data & Comments
Data

 Cite This Article 2 clicks
Cite This Article 2 clicks


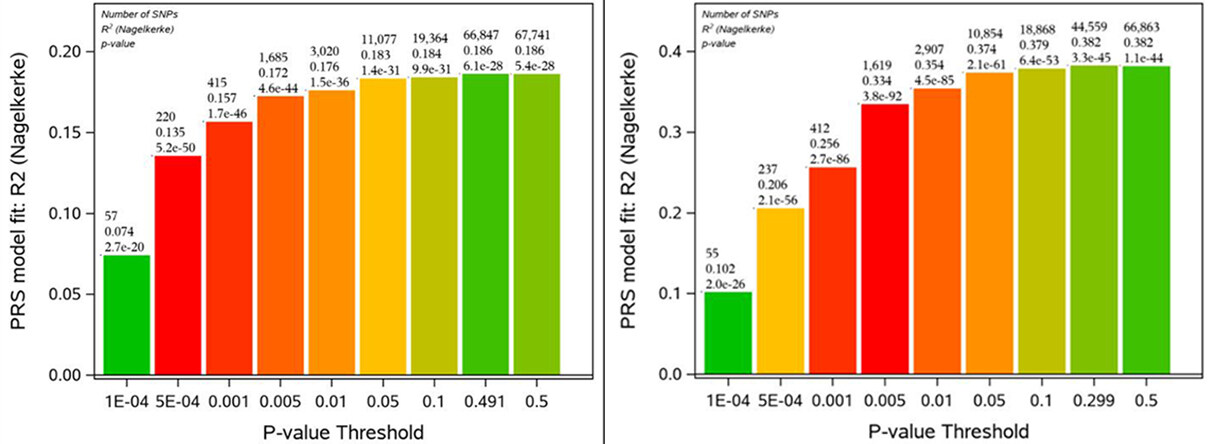








Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.