Retinal organoid differentiation methods determine organoid cellular composition
Abstract
Aim: We aimed to compare the quantity and quality of aging retinal organoids generated by applying three distinct differentiation protocols for human-derived induced pluripotent stem cells (hiPSC).
Methods: hiPSC were differentiated to retinal organoids using a 3D technique (Method 1) and a 3D-2D-3D technique (Method 2), the latter modified by the addition of BMP4 (Method 3). To investigate the retinal organoid quantity, we counted the number of retinal domains, precursors to organoids during differentiation. The retinal organoid quality was evaluated by immunostaining for markers of different retinal cell types in whole cryosections after days 85, 120, and 200 in culture.
Results: Method 3 produced strikingly more retinal domains per differentiation (65 ± 27) than Methods 1 (12.3 ± 11.2) and 2 (6.3 ± 6.7). Furthermore, retinal organoids differentiated with Method 3 contained significantly more CRX-positive photoreceptors and BRN3A-positive ganglion cells after 85 days in culture, compared to Methods 1 and 2. After 200 days in culture, the retinal organoids differentiated with Method 3 showed proper maturation, as demonstrated by the expression of mature rod and cone photoreceptor markers.
Conclusion: This study demonstrates that the retinal organoid differentiation method can significantly impact the cellular composition of retinal organoids at various time points of development.
Keywords
INTRODUCTION
Organoids are three-dimensional (3D) in vitro miniature organs, which contain multiple organ-specific cell types and a comparable spatial organization to the native tissue[1]. To date, differentiation protocols for multiple organoid types exist, including cerebral organoids[2-5], intestinal organoids[6,7], lung organoids[8,9], and kidney organoids[10], among many others. Organoid research was pioneered in 2005 when the Sasai group developed a protocol to selectively differentiate murine embryonic stem cells to neurons[2,11]. In the following years, the same group developed methods to differentiate murine and later human embryonic stem cells to optic cups[12,13]. These advances served as a cornerstone for other groups to develop human induced pluripotent stem cell (hiPSC) derived retinal organoid differentiation protocols[14-16].
Retinal organoids provide many exciting new avenues of research, in part filling a void created by the incongruencies between animal models and human diseases. hiPSC derived retinal organoids contain the main cell types native to the retina: rod and cone photoreceptors, bipolar cells, horizontal cells, amacrine cells, ganglion cells, and Müller cells. The development and molecular profile of the organoid cells appear to be fairly similar to their native counterparts[17,18]. Recently, retinal organoids have also been used to model inherited retinal dystrophies[19-23], although it seems that organoids may be best suited to model severe phenotypes with an early disease onset.
To date, a variety of retinal organoid differentiation protocols have been established. Broadly, the procedures can be classified into two main categories: 3D protocols and 3D-2D-3D protocols[24]. The 3D differentiation protocols are based on the serum-free embryoid body-like quick aggregation culture technique developed by Eiraku et al.[2,11] and Watanabe et al.[25]. In this technique, the cells are cultured in suspension for the entire differentiation process (referred to as “3D” culture). Later, other groups added a temporary adherent phase (the “2D” phase) after the aggregation step, which improved the retinal domain development[26]. Retinal domains are organoid precursors, which are excised and cultured in suspension until they are harvested (the second “3D” phase)[26]. Protocols from both main categories can be modified by including a variety of possible extrinsic chemical signals to promote retinal differentiation or enhance cell survival.
In this study, the efficacy of differentiation protocols from both main categories was investigated including a protocol by Wahlin et al.[15] which operates according to the 3D technique (Method 1). This differentiation protocol utilizes a Wnt inhibitor (Inhibitor of Wnt response compound-1-endo, IWR-1e) to induce retinal differentiation[27], a Hedgehog agonist [Smoothened agonist (SAG)] to enhance the survival of neural cells[28], and a Notch signaling inhibitor {N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester, DAPT} to increase photoreceptor yield[29]. The differentiation protocol by Zhong et al.[14] is based on the 3D-2D-3D technique (Method 2) and uses minimal extrinsic inductive cues to facilitate autonomous differentiation. We also tested an adaptation of Method 2 which includes a brief early exposure to bone morphogenic protein 4 (BMP4), as developed by Kuwahara et al.[16] and more recently demonstrated by Capowski et al.[30] (Method 3). The overall aim of this study was to compare quantity and quality of retinal organoids produced by the three methods and to identify any discrepancies in cellular composition upon long-term development.
METHODS
hiPSC reprogramming and maintenance
The isolation of adult human dermal fibroblasts and reprogramming to hiPSC (approved by the local ethics committee reference No. 11-101-0228) were performed according to the method in[31] with modifications as per the work in[32] with minor additional modifications. Briefly, hiPSCs were reprogrammed via polycistronic lentiviral transduction utilizing the Human STEMCCA Cre-Excisable Constitutive Polycistronic (OKS/L-Myc) Lentivirus Reprogramming Kit (Sigma-Aldrich, St. Louis, USA) according to the manufacturer’s instructions. hiPSCs were cultured in mTeSR™ Plus medium (Stemcell Technologies, Cologne, Germany) on plates coated with hESC-qualified Matrigel (Corning®, New York, USA) and cryopreserved in CryoStor® CS10 medium (Stemcell Technologies, Cologne, Germany).
Retinal organoid differentiation
Method 1
Retinal organoids were differentiated according to Wahlin et al.[15] with minor modifications. All cells were cultured under normoxia. On day 0, hiPSCs were treated with StemPro™ Accutase™ (Life Technologies, California, USA) for 9 min, and 2000 cells per well were plated on a Corning® Costar® Ultra-Low Attachment 96 Well Plate (Sigma-Aldrich, St. Louis, USA) in mTeSR™ Plus medium (Stemcell Technologies, Cologne, Germany) with 5 µm Blebbistatin (Cayman Chemical, Michigan, USA). Cells were weaned onto BE6.2 medium [Supplementary Tables 1 and 2]. Then, 3 µM IWR-1e (Sigma-Aldrich, St. Louis, USA) was added to the medium until day 7, and 1% Matrigel Growth Factor Reduced Basement Membrane Matrix (Corning®, New York, USA) was added until day 8.
On day 10, the aggregates which formed retinal domains were counted and isolated with 27G cannulas (BD, Franklin Lakes, USA) attached to 1 mL syringes (BD, Franklin Lakes, USA). Thereafter, the retinal organoids were cultured in separate wells of Corning™ Costar™ Ultra-Low Attachment 24 Well Plates (Thermo Fisher Scientific, Waltham, USA) following Hallam et al.[33]. From day 10 to day 18, 100 nM SAG (Cayman Chemical, Michigan, USA) was added to the medium. On day 12, the medium was switched to LTR medium [Supplementary Table 3]. After day 20, 500 nM all-trans retinoic acid (Cayman Chemical, Michigan, USA) and, from day 29 to day 42, 10 µM DAPT (Cayman Chemical, Michigan, USA) were added to the medium.
Method 2
Retinal organoids were differentiated according to Zhong et al.[14] with minor modifications. On day 0, hiPSCs were treated with Dispase (Stemcell Technologies, Cologne, Germany) for 15 min and transferred to Corning® Ultra-Low attachment cell culture flasks (Sigma-Aldrich, St. Louis, USA) in mTeSR™ Plus medium (Stemcell Technologies, Cologne, Germany) with 10 µm Blebbistatin (Cayman Chemical, Michigan, USA) to facilitate aggregate formation. Over the course of three days, the aggregates were weaned onto NIM medium [Supplementary Table 4]. A medium exchange was performed on day 6, and, on day 7, the aggregates were plated on Matrigel Growth Factor Reduced Basement Membrane Matrix (Corning®, New York, USA) coated CELLSTAR® Cell Culture 6 Well Plates (Greiner Bio-One, Kremsmuenster, Austria). Half the medium was exchanged for fresh NIM every three days. On day 16, the medium was switched to RDM medium [Supplementary Table 5]. On day 23, the retinal domains were counted. Then, 10 mM HEPES (Thermo Fisher Scientific, Waltham, USA) was added to the medium, and the retinal organoids were isolated as described in Method 1 and transferred to separate wells of a Corning® Costar® Ultra-Low Attachment 96 Well Plate (Sigma-Aldrich, St. Louis, USA) following Hallam et al.[33]. On day 43, the medium was switched to RC2 medium [Supplementary Table 6]. From day 63 to day 90, 1 µM all-trans retinoic acid (Cayman Chemical, Michigan, USA) was added to the medium five times weekly. On day 91, the medium was switched to RC1 medium [Supplementary Table 7].
Method 3
Retinal organoids were differentiated as described in Method 2, with a single modification. On day 6, 1.5 nM BMP4 was added to the medium, as demonstrated by Capowski et al.[30].
Immunofluorescence
Immunocytochemistry
Cryosections (10 µm) of retinal organoids were washed three times in PBS for 5 min at room temperature (RT) and blocked in 10% normal goat serum (Abcam, Cambridge, UK) and 0.3% Triton X (VWR, Pennsylvania, USA) in PBS for 1 h at RT. Samples were incubated in primary antibodies in 2.5% normal goat serum and 0.1% Triton X in PBS overnight (ON) at 8 °C. Samples were washed three times in PBS for 5 min at RT, and incubated in the secondary antibodies in 2.5% normal goat serum, 0.5% 4’,6-diamidino-2-phenylindole (DAPI, Molecular Probes, Leiden, the Netherlands), and 0.1% Triton X in PBS ON at 8 °C. The secondary antibody solutions were aliquoted and stored at -20 °C, to reduce batch effects. Samples were washed three times in PBS for 5 min at room temperature and mounted in Dako fluorescence mounting medium (Agilent, California, USA). Slides were dried ON before imaging.
Antibodies
The following primary antibody dilutions were used: AP2A 1:250 (ab108311, Abcam, Cambridge, UK), BRN3A 1:250 (ab245230, Abcam, Cambridge, UK), CRX 1:1000 (H00001406-M02, Abnova, Taipeh, Taiwan), OPN1SW 1:1000 (JH455, gift from Jeremy Nathans, Johns Hopkins University, Maryland, USA), RCVRN 1:1000 (AB5585, Sigma, Missouri, USA), RHO1D4 1:1000 (gift from Robert S. Molday, University of British Columbia, Vancouver, Canada), SNCG 1:500 (H00006623-M01, Abnova, Taipeh, Taiwan), and VSX2 1:1000 (HPA003436, Sigma, Missouri, USA). The following secondary antibodies and dilutions were used: Alexa Fluor® 488 goat anti-rabbit IgG 1:667 (Thermo Fisher Scientific, Waltham, USA) and Alexa Fluor® 594 goat anti-mouse IgG 1:667 (Thermo Fisher Scientific, Waltham, USA). For samples stained with PNA, Alexa Fluor® 594 goat anti-mouse IgG was replaced with 4% Alexa 594-conjugated peanut agglutinin (Invitrogen, California, USA).
Analysis
Retinal organoid yield quantification
The number of retinal domains was manually counted on day 10 for Method 1 and day 23 for Methods 2 and 3. The mean from three independent differentiations was calculated. To test for significance, a one-way ANOVA test, post hoc Tukey test, and Bonferroni correction for multiple comparisons were performed.
The number of retinal organoids was counted on day 63 for Methods 2 and 3, and the mean was calculated from three independent differentiations. A Mann-Whitney U-test was performed to test for significance.
Microscopy and image analysis
All slides were imaged on a confocal microscope Olympus Fluoview FV3000. To confirm successful retinal organoid differentiation and maturation, closeup images of retinal organoid sections were taken. To control for regional variability, the quantification of the relative expression of retinal markers was performed from whole cryosections. Three organoids were analyzed per method and each timepoint. The relative area of the stain was calculated compared to the DAPI-positive area (for nuclear markers AP2A, BRN3A, CRX, and SNCG) or the whole cryosection (for cytosolic markers RCVRN and RHO). The quantification was performed in Fiji, an updated distribution of ImageJ[34]. Macros were written for the quantification, which converted the images to 16 bits, performed thresholding according to method in[35], and measured the remaining area.
Statistical analysis
The Kolmogorov-Smirnov test was used to test for normality. Significance was investigated as per the retinal organoid yield quantification. To compare organoids from two timepoints, a Student’s t-test was performed, and a P-value < 0.05 was considered significant.
RESULTS
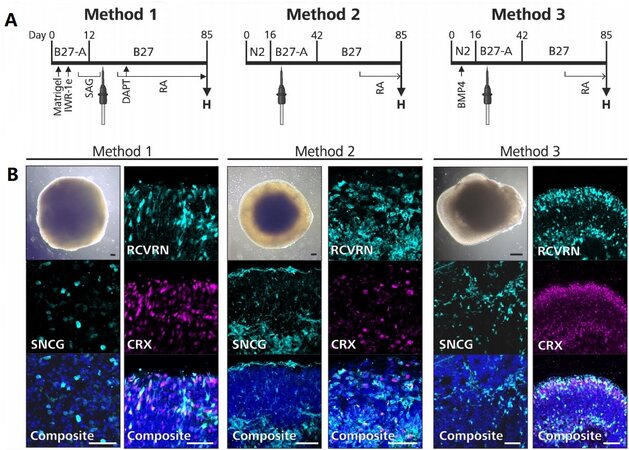
To investigate the efficacy of retinal organoid differentiation protocols, we differentiated the same hiPSC line to retinal organoids using three differentiation protocols termed Method 1 (based on Wahlin et al.[15]), Method 2 (based on Zhong et al.[14]), and Method 3 (based on Capowski et al.[30]) [Figure 1A]. First, we investigated whether each differentiation method could successfully generate retinal organoids. In our hands, each differentiation method successfully produced retinal organoids showing a phase bright outer rim, which corresponds to the developing neuroepithelium[14,15,30], known as a characteristic morphological feature of high-quality retinal organoids [Figure 1B]. To further confirm effective retinal differentiation, the expression of a ganglion[36,37] and amacrine cell[38] marker synuclein gamma (SNCG) and two early photoreceptor markers recoverin (RCVRN)[39] and cone-rod homeobox (CRX)[40,41] were confirmed in retinal organoids from each differentiation method [Figure 1B]. We used these stains to evaluate the histoarchitecture of the retinal organoids and observed an optimal lamination in the organoids from Method 3.
Figure 1. Overview of retinal organoid differentiation protocols. (A) A timeline of each retinal organoid differentiation protocol is shown. The supplements used in each protocol are denoted above the timeline, the extrinsic factors are denoted below the timeline, and the timepoint of excision is indicated with a tungsten needle. “H” indicates the timepoint when the retinal organoids were harvested for the comparative analyses. (B) Each differentiation method successfully produced retinal organoids (shown on day 85). Retinal organoids from each method contained amacrine and ganglion cells (SNCG-positive) and juvenile photoreceptor cells (RCVRN-positive, CRX-positive). Composite images were counterstained with DAPI. IWR-1e: Inhibitor of Wnt response compound-1-endo; SAG: smoothened agonist; DAPT: N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester; RA: all-trans retinoic acid; H: harvest; BMP4: bone morphogenic protein 4; SNCG: synuclein gamma; RCVRN: recoverin; CRX: cone-rod homeobox. Black scale bar: 100 µm; white scale bar: 50 µm.
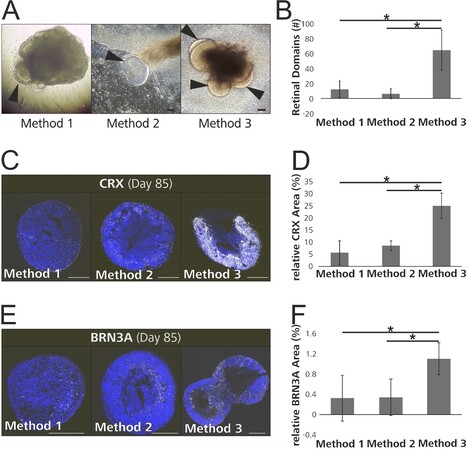
The primary objective of this study was to identify the differentiation method which produced the highest yield and best quality of retinal organoids. To determine the retinal organoid quantity, we counted the retinal domains produced using Method 1 on day 10 and using Methods 2 and 3 on day 23 [Figure 2A and B]. These timepoints were chosen based on recommendations given by Wahlin et al.[15] and Zhong et al.[14] concerning the optimal timepoint for retinal domain excision. Method 3 produced strikingly more retinal domains per differentiation (65 ± 27) than Method 1 (12.3 ± 11.2) and Method 2 (6.3 ± 6.7), despite some variability. Furthermore, the retinal domains from Method 3 were more clearly defined, and therefore easier to excise [Figure 2A]. The number of retinal domains which matured to retinal organoids was counted on day 63. Again, Method 3 produced more retinal organoids than Method 2 (24.7 ± 17.2 vs. 1.7 ± 0.6)
Figure 2. Method 3 produced a larger quantity and higher quality of retinal organoids. (A) Brightfield images show proper differentiation of retinal domains pre-excision (arrowheads indicate retinal domains). (B) More organoids per differentiation were acquired using Method 3 compared to Method 2. (C) Whole cryosections of 85-day-old retinal organoids from each differentiation method, stained with an early photoreceptor marker (CRX) and counterstained with DAPI are shown. (D) The relative CRX-positive area was greatest in the retinal organoids differentiated using Method 3. (E) Whole cryosections of 85-day-old retinal organoids stained with a ganglion cell marker (BRN3A) and counterstained with DAPI are shown. (F) The relative BRN3A positive area was greatest in the retinal organoids differentiated using Method 3. CRX: Cone-rod homeobox; BRN3A: POU class 4 homeobox. Scale bar (A) 100 µm; (C, E) 300 µm. (B, D, F) *P < 0.017 (Bonferroni-corrected).
To investigate the quality of retinal organoids produced by the three different methods, we quantified the expression of several retinal markers in 85-day-old retinal organoids. This timepoint was chosen, since the cellular composition of retinal organoids at this timepoint has been well documented by previous groups[14,30,42]. To control for regional variability, we analyzed whole cryosections. First, we investigated the expression of CRX, a transcription factor which is one of the earliest genes expressed in photoreceptor precursors, as well as mature photoreceptors[40,41,43]. Retinal organoid cryosections were immunostained for CRX [Figure 2C], and the CRX-positive area was quantified relative to the DAPI-positive area (blue staining). We found the highest CRX expression in the retinal organoids differentiated using Method 3 (25% ± 5.3%), while the CRX expressions for Methods 1 (5.7% ± 4.8%) and 2 (8.6% ± 2%) were significantly lower than that of Method 3 but comparable to each other [Figure 2D]. Next, we investigated the expression of BRN3A, a transcription factor and an early retinal ganglion cell marker[44-46] [Figure 2E]. Again, the retinal organoids produced by Method 3 showed the highest BRN3A expression (Method 1: 0.3% ± 0.5%; Method 2: 0.3% ± 0.4%; Method 3: 1.8% ± 0.3%) [Figure 2F]. In contrast, the different methods did not have a significant effect on the expression of the amacrine cell marker transcription factor AP-2-alpha, the ganglion cell marker SNCG, and the photoreceptor marker RCVRN [Supplementary Figure 2].
To more conclusively investigate whether the retinal organoids generated with Method 3 differentiate properly, we performed multiple analyses of long-term cultures. Again, the timepoints were chosen based on the availability of reference data from previous publications[14,30]. We first investigated the RCVRN-positive area relative to the whole cryosection, in 85- and 120-day-old retinal organoids [Figure 3A]. The relative RCVRN-positive area was significantly higher in the 120-day-old retinal organoids, indicating more progenitor cells differentiated along a photoreceptor lineage over time [Figure 3B]. Next, the relative expression of RHO, a mature rod photoreceptor marker, was analyzed in 120- and 200-day-old organoids [Figure 3C]. Again, we observed that the expression of this marker increased over time, indicating that the juvenile photoreceptors in the organoids mature to rods over time [Figure 3D]. Finally, we examined the localization of visual system homeobox 2 (VSX2) expression in 200-day-old organoids. During retinal development, VSX2 is initially expressed by neural retinal progenitor cells and later is restricted to bipolar cells in the developing inner nuclear layer (INL)[14,47]. We localized VSX2 in the developing inner nuclear layer in 200-day-old retinal organoids [Figure 3E]. In addition, OPN1SW+ blue cone photoreceptors and PNA positive cone photoreceptor inner and outer segments[48] were identified in the 200-day-old retinal organoid [Figure 3F].
Figure 3. Retinal organoids differentiated by following Method 3 acquire mature characteristics. (A) Retinal organoids from two timepoints, stained with a developing photoreceptor marker (RCVRN) and counterstained with DAPI are shown. (B) The relative RCVRN-positive area was greater in 120-day-old retinal organoids compared to 85-day-old retinal organoids. (C) Retinal organoids from two timepoints, stained with a mature rod photoreceptor marker (RHO) and counterstained with DAPI are shown. (D) The relative RHO-positive area was larger in the retinal organoids harvested on 200-day-old retinal organoids compared to 120-day-old retinal organoids. (E) VSX2 expression in the outer nuclear layer of an 85-day-old retinal organoid, counterstained with DAPI. (F) VSX2 expression in the inner nuclear layer of a 200-day-old retinal organoid, counterstained with DAPI. (G) A 200-day old retinal organoid showed surface projections. (H) The retinal organoid shown in (G) displayed PNA expression in the putative inner and outer segments and contained OPN1SW-positive blue cone photoreceptors. Counterstaining was performed with DAPI. RCVRN: Recoverin; RHO: rhodopsin; ONL: outer nuclear layer; INL: inner nuclear layer; VSX2: visual system homeobox 2; PNA: peanut agglutinin; OPN1SW: opsin 1, short wave sensitive. Scale bar: (A, C) 300 µm; (E, F) 50 µm; (G) 1 mm; (H) 50 µm. *P < 0.05, **P < 0.01.
DISCUSSION
In this study, we differentiated hiPSCs to retinal organoids following the original descriptions of three protocols which implement different techniques and extrinsic differentiation stimuli. The starting point (iPSC) and endpoint (retinal organoids) of the three methods are identical, but otherwise the methods are very different. We chose to adhere to the published protocols instead of manipulating them to reduce their dissimilarities, in order to compare the original protocols. While each differentiation protocol successfully generated retinal organoids, Method 3 produced significantly more organoids per differentiation, and the organoids contained more CRX-positive photoreceptors and BRN3A-positive ganglion cells after 85 days in culture. We further investigated 200-day-old retinal organoids differentiated based on Method 3 and observed proper maturation indicated by the increase of RCVRN- and RHO-positive cells, VSX2 expression in the INL, and OPN1SW and PNA expression in the putative inner and outer segments. Our comparison demonstrates that Method 3 produced the highest quantity and quality of retinal organoids. Interestingly, Method 3 only increased the proportion of BRN3A- and CRX-positive cells and not the proportion of AP-2-alpha-, SNCG-, or RCVRN-positive cells. Each of these markers is expressed by a different subset of retinal cells, and some markers are not exclusive to one cell type. For example, although BRN3A[44-46] and SNCG[36,37] are both expressed in ganglion cells, there is evidence that some amacrine cells also express SNCG[38]. The AP-2-alpha amacrine cell marker which we investigated in this study was not significantly altered as a consequence of the differentiation method. Taken together, these results indicate that Method 3 increases the proportion of ganglion cells but not amacrine cells in the retinal organoids. This underscores that all three methods successfully generated retinal organoids, while the differentiation method substantially alters their cellular composition. This is especially interesting when comparing Methods 2 and 3, since only a single extrinsic inductive stimulus (BMP4) significantly altered the cellular composition of the ensuing retinal organoids.
BMP4 belongs to a family of signaling molecules which were initially discovered for their ability to induce bone formation[49] but are now known to play a number of crucial roles in other organ systems[50]. Germline deletion of BMP4 is embryologically lethal in mice between 6.5 and 9.5 days post conception[51]. Conditional deletion of BMP4 from the optic vesicle resulted in an optic cup devoid of neural retinal markers, indicating that BMP4 is required for retinal specification[52]. In vitro, BMP4 does not appear to be required for retinal differentiation, since the protocols without BMP4 also produce organoids. Instead, it is possible that BMP4 exposure reinforces or amplifies retinal differentiation in vitro, while the timepoint of BMP4 exposure is thought to be critical[53]. An effect of the differentiation method on the ensuing organoid has previously been observed in the study of cortical organoids. Tanaka et al.[54] performed a synthetic comparative analysis of single-cell transcriptome profiling of cortical organoids from eight different protocols and observed that the cellular composition and differentiation routes varied across the individual differentiation protocols. In line with these findings, Chichagova et al.[55] compared two retinal organoid differentiation methods and found that, with one of their hiPSC lines, one method was skewed towards an interneuron fate, whereas another method skewed the retinal organoids towards a rod photoreceptor fate.
Ours and similar studies which compare retinal organoid differentiation protocols are vital for the scientific community, especially since there are still some open questions. Based on our study, we should not surmise that 3D-2D-3D techniques (such as Methods 2 and 3) are inherently superior to 3D techniques (such as Method 1). In our hands, the organoids from Method 1 showed insufficient lamination despite staining positive for several retinal cell types. Notably, the lamination in the original publication is above reproach. It is rather speculative to question why Method 1 did not work better in our hands, especially because the method was described very well in the original publication. Furthermore, retinal organoids are plagued by a certain degree of variability[33]. In our study, the quantification of the retinal domains showed some variability, which may have been caused by small numbers and differences in the counting methodology. It is plausible that weaker effects may have been masked by the variability.
To date, many retinal organoid differentiation protocols[18,42,56,57] have not been tested in side-to-side comparisons by independent groups, most likely due to the considerable time commitment associated with these protocols. To facilitate the comparability of experimental applications of retinal organoids across studies, standardized retinal organoid differentiation protocols are needed, which should focus on the type of differentiation technique and the extrinsic differentiation cues.
DECLARATIONS
AcknowledgmentsWe sincerely thank Prof. M. Valeria Canto-Soler (Department of Ophthalmology, University of Colorado School of Medicine) for providing us with a detailed differentiation protocol. We would also like to thank Nico Hertl for his valuable help and advice in hiPSC maintenance.
Authors’ contributionsDesigned the study, performed data acquisition, analysis and interpretation, prepared the manuscript: Berber P
Made substantial contributions to the discussion of content, provided feedback and performed manuscript editing: Milenkovic A
Made substantial contributions to method optimization, provided excellent technical support: Michaelis L
Conceptualized and supervised the study, helped with data interpretation and condensed presentation in Tables and Figures, performed manuscript editing and finalization: Weber BHF
Made final approval of the manuscript: Milenkovic A, Michaelis L, Weber BHF
Availability of data and materialsNot applicable.
Financial support and sponsorshipThe study was supported in parts by a grant from the German Research Foundation (DFG) to BHFW (WE1259/32-1) and institutional funds of the Institute of Human Genetics (Title 77). The funding body had no role in the experimental design, collection, analysis and interpretation of data, and writing of the manuscript. The FV3000 confocal microscope was funded by a grant (INST 89/506-1 FUGG, 91b GG) from the Deutsche Forschungsgemeinschaft (DFG).
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateThe reprogramming and differentiation of hiPSC from human donor material were approved by the local ethics review board of the University of Regensburg, Germany (reference No. 11-101-0228) and have been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. Informed consent was given by the proband participating in the study.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
1. Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014;345:1247125.
2. Eiraku M, Watanabe K, Matsuo-Takasaki M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008;3:519-32.
3. Lancaster MA, Corsini NS, Wolfinger S, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol 2017;35:659-66.
4. Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature 2013;501:373-9.
5. Velasco S, Kedaigle AJ, Simmons SK, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 2019;570:523-7.
6. Schwank G, Koo BK, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013;13:653-8.
7. Dekkers JF, Wiegerinck CL, de Jonge HR, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 2013;19:939-45.
8. Longmire TA, Ikonomou L, Hawkins F, et al. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 2012;10:398-411.
9. Wong AP, Bear CE, Chin S, et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat Biotechnol 2012;30:876-82.
10. Low JH, Li P, Chew EGY, et al. Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell 2019;25:373-87.e9.
11. Watanabe K, Kamiya D, Nishiyama A, et al. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci 2005;8:288-96.
12. Eiraku M, Takata N, Ishibashi H, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011;472:51-6.
13. Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012;10:771-85.
14. Zhong X, Gutierrez C, Xue T, et al. Generation of three dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 2014;5:4047.
15. Wahlin KJ, Maruotti JA, Sripathi SR, et al. Photoreceptor outer segment-like structures in long-term 3D retinas from human pluripotent stem cells. Sci Rep 2017;7:766.
16. Kuwahara A, Ozone C, Nakano T, Saito K, Eiraku M, Sasai Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat Commun 2015;6:6286.
17. Sridhar A, Hoshino A, Finkbeiner CR, et al. Single-cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures. Cell Rep 2020;30:1644-59.e4.
18. Cowan CS, Renner M, De Gennaro M, et al. Cell Types of the human retina and its organoids at single-cell resolution. Cell 2020;182:1623-40.e34.
19. Lane A, Jovanovic K, Shortall C, et al. Modelling and rescue of RP2 retinitis pigmentosa using iPSC derived retinal organoids. Stem Cell Reports 2020;15:67-79.
20. Huang KC, Wang ML, Chen SJ, et al. Morphological and molecular defects in human three-dimensional retinal organoid model of X-linked juvenile retinoschisis. Stem Cell Reports 2019;13:906-23.
21. Kruczek K, Qu Z, Gentry J, et al. Gene Therapy of Dominant CRX-Leber Congenital Amaurosis using Patient Stem Cell-Derived Retinal Organoids. Stem Cell Reports 2021;16:252-63.
22. Gao ML, Lei XL, Han F, et al. Patient-specific retinal organoids recapitulate disease features of late-onset retinitis pigmentosa. Front Cell Dev Biol 2020;8:128.
23. Quinn PM, Buck TM, Mulder AA, et al. Human iPSC-derived retinas recapitulate the fetal CRB1 CRB2 complex formation and demonstrate that photoreceptors and müller glia are targets of AAV5. Stem Cell Reports 2019;12:906-19.
24. Kruczek K, Swaroop A. Pluripotent stem cell-derived retinal organoids for disease modeling and development of therapies. Stem Cells 2020;38:1206-15.
25. Watanabe K, Ueno M, Kamiya D, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 2007;25:681-6.
26. Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci U S A 2009;106:16698-703.
27. Zhou S, Flamier A, Abdouh M, et al. Differentiation of human embryonic stem cells into cone photoreceptors through simultaneous inhibition of BMP, TGFβ and Wnt signaling. Development 2015;142:3294-306.
28. Bragina O, Sergejeva S, Serg M, et al. Smoothened agonist augments proliferation and survival of neural cells. Neurosci Lett 2010;482:81-5.
29. Osakada F, Ikeda H, Mandai M, et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol 2008;26:215-24.
30. Capowski EE, Samimi K, Mayerl SJ, et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 2019;146:dev171686.
31. Brandl C, Zimmermann SJ, Milenkovic VM, et al. In-depth characterisation of Retinal Pigment Epithelium (RPE) cells derived from human induced pluripotent stem cells (hiPSC). Neuromolecular Med 2014;16:551-64.
32. Nachtigal AL, Milenkovic A, Brandl C, et al. Mutation-dependent pathomechanisms determine the phenotype in the bestrophinopathies. Int J Mol Sci 2020;21:1597.
33. Hallam D, Hilgen G, Dorgau B, et al. Human-induced pluripotent stem cells generate light responsive retinal organoids with variable and nutrient-dependent efficiency. Stem Cells 2018;36:1535-51.
34. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676-82.
35. Huang L, Wang MJ. Image thresholding by minimizing the measures of fuzziness. Pattern Recognition 1995;28:41-51.
36. Lukowski SW, Lo CY, Sharov AA, et al. A single-cell transcriptome atlas of the adult human retina. EMBO J 2019;38:e100811.
37. Soto I, Oglesby E, Buckingham BP, et al. Retinal ganglion cells downregulate gene expression and lose their axons within the optic nerve head in a mouse glaucoma model. J Neurosci 2008;28:548-61.
38. Kunzevitzky NJ, Almeida MV, Goldberg JL. Amacrine cell gene expression and survival signaling: differences from neighboring retinal ganglion cells. Invest Ophthalmol Vis Sci 2010;51:3800-12.
39. Yan XX, Wiechmann AF. Early expression of recoverin in a unique population of neurons in the human retina. Anat Embryol (Berl) 1997;195:51-63.
40. Furukawa T, Morrow EM, Cepko CL. Crx, a Novel otx-like Homeobox Gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 1997;91:531-41.
41. Chen S, Wang Q, Nie Z, et al. Crx, a Novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 1997;19:1017-30.
42. Gonzalez-Cordero A, Kruczek K, Naeem A, et al. Recapitulation of human retinal development from human pluripotent stem cells generates transplantable populations of cone photoreceptors. Stem Cell Reports 2017;9:820-37.
43. Muranishi Y, Sato S, Inoue T, et al. Gene expression analysis of embryonic photoreceptor precursor cells using BAC-Crx-EGFP transgenic mouse. Biochem Biophys Res Commun 2010;392:317-22.
44. Xiang M, Zhou L, Macke J, et al. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci 1995;15:4762-85.
45. Sajgo S, Ghinia MG, Brooks M, et al. Molecular codes for cell type specification in Brn3 retinal ganglion cells. Proc Natl Acad Sci U S A 2017;114:E3974-83.
46. Luo Z, Xu C, Li K, et al. Islet1 and Brn3 Expression Pattern Study in Human Retina and hiPSC-Derived Retinal Organoid. Stem Cells Int 2019;2019:8786396.
47. Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A 1996;93:589-95.
48. Blanks JC, Johnson L v. Specific binding of peanut lectin to a class of retinal photoreceptor cells. A species comparison. Invest Ophthalmol Vis Sci 1984;25:546-57.
50. Wang RN, Green J, Wang Z, et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis 2014;1:87-105.
51. Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev 1995;9:2105-16.
52. Huang J, Liu Y, Oltean A, Beebe DC. Bmp4 from the optic vesicle specifies murine retina formation. Dev Biol 2015;402:119-26.
53. Wagstaff PE, Heredero Berzal A, Boon CJF, Quinn PMJ, Ten Asbroek ALMA, Bergen AA. The role of small molecules and their effect on the molecular mechanisms of early retinal organoid development. Int J Mol Sci 2021;22:7081.
54. Tanaka Y, Cakir B, Xiang Y, Sullivan GJ, Park IH. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep 2020;30:1682-9.e3.
55. Chichagova V, Hilgen G, Ghareeb A, et al. Human iPSC differentiation to retinal organoids in response to IGF1 and BMP4 activation is line- and method-dependent. Stem Cells 2020;38:195-201.
56. Reichman S, Terray A, Slembrouck A, et al. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc Natl Acad Sci U S A 2014;111:8518-23.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Berber P, Milenkovic A, Michaelis L, Weber BHF. Retinal organoid differentiation methods determine organoid cellular composition. J Transl Genet Genom 2021;5:292-303. http://dx.doi.org/10.20517/jtgg.2021.35
AMA Style
Berber P, Milenkovic A, Michaelis L, Weber BHF. Retinal organoid differentiation methods determine organoid cellular composition. Journal of Translational Genetics and Genomics. 2021; 5(3): 292-303. http://dx.doi.org/10.20517/jtgg.2021.35
Chicago/Turabian Style
Berber, Patricia, Andrea Milenkovic, Lisa Michaelis, Bernhard H. F. Weber. 2021. "Retinal organoid differentiation methods determine organoid cellular composition" Journal of Translational Genetics and Genomics. 5, no.3: 292-303. http://dx.doi.org/10.20517/jtgg.2021.35
ACS Style
Berber, P.; Milenkovic A.; Michaelis L.; Weber BHF. Retinal organoid differentiation methods determine organoid cellular composition. J. Transl. Genet. Genom. 2021, 5, 292-303. http://dx.doi.org/10.20517/jtgg.2021.35
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 46 clicks
Cite This Article 46 clicks












Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.