Silent players, loud impact: unveiling the therapeutic potentials of LncRNAs
Abstract
Long non-coding RNAs (lncRNAs) are a class of RNA transcripts that are long (i.e., more than 200 nucleotides) and not translated into proteins. They have recently emerged as potential treatment targets for numerous disorders due to their involvement in multiple cellular functions such as gene regulation, epigenetic modulation, and chromatin organization. This review highlights the current state of lncRNA-based therapeutics, the potential of lncRNAs as drug targets for treating human diseases, the various strategies and types of RNA-based therapeutics, and the complications of developing lncRNA-based drugs. We conclude that lncRNA-based therapeutics represent a promising class of drugs that can potentially treat various human diseases and that further research is needed to fully realize their therapeutic potential.
Keywords
INTRODUCTION
Studies have shown that only 2% of the genome consists of protein-coding genes[1], and the remaining 98% do not have protein-encoding function[2]. Surprisingly, most of the genome (around 70%) is actively transcribed, meaning that most of the human transcribed RNAs consist of non-coding RNAs (ncRNAs), genes that are transcribed into RNA but not translated into proteins[1]. According to their size, the ncRNAs are divided into two groups: small ncRNAs (< 200 nt) and long ncRNAs (LncRNAs)[3]. The category of small ncRNAs encompasses a diverse range of RNA types, including microRNAs, piwiRNAs, small nucleolar RNAs[4], and small interfering RNAs (siRNAs)[5]. lncRNAs are a class of RNA transcripts that are long (i.e., more than 200 nucleotides) and not translated into proteins[6].
Small ncRNAs have been the subject of extensive research for a long time due to their significant functions in the initiation and prognosis of many diseases. On the other hand, lncRNAs are not well understood compared to other classes of ncRNAs[5], despite the fact that lncRNAs account for more than 80% of ncRNAs[2]. LncRNAs often exhibit low expression levels and are typically associated with specific tissues or cell types[7]. They have been growing as new contributors to cancer in both oncogenic and tumor suppressive pathways[8]. They have also recently been identified as significant players in both regular cellular functions and disease development[9]. Additionally, lncRNAs are crucial in numerous physiological processes, including RNA splicing and X-chromosome inactivation[10].
CLASSIFICATION
The classification of lncRNAs, based on both their transcriptional orientation and their proximity to adjacent protein-coding genes, encompasses four main types: Intergenic, Bidirectional, Antisense, and Sense lncRNAs. Intergenic lncRNAs hold a significant position within human lncRNAs, constituting the largest subgroup. Positioned in genomic regions without protein-coding sequences within a five-kilobase range[11], intergenic lncRNAs act as genomic separators between two protein-coding genes[5]. Bidirectional lncRNAs, another distinct type, are transcribed from the promoter of protein-coding genes but in the opposite direction[12]. Antisense lncRNAs form the second most prevalent group in humans[5]. They are transcribed from the antisense strand of protein-coding genes, often overlapping with exonic or intronic regions. Finally, Sense lncRNAs, a unique category, originate from the sense strand of protein-coding genes. This type includes instances where the lncRNA either partially overlaps with the gene, including portions of exons, or entirely encompasses the gene's sequence within an intron[4]. This diverse classification is depicted in Figure 1.
Figure 1. Classification of lncRNAs. Republished with permission from[12].
FUNCTION
The functions of lncRNAs, like protein-coding genes, are diverse and vary greatly[1]. LncRNAs have been found to play a role in various significant biological processes via their interaction with DNA or chromatin, their effect on regulatory or signaling pathways, and their regulatory function over other types of RNAs[5]. LncRNAs may function as decoys that can attract and prevent effector proteins from carrying out their functions via binding and diverting them from their intended target site[6]. Some lncRNAs can have microRNA response elements, which are complementary sites for microRNAs. These are called competitive endogenous RNAs, acting as molecular sponges for microRNAs, inhibiting them from reaching the target genes of interest[11].
LncRNAs have several ways to regulate gene expression, with the most frequent being an epigenetic process that frequently causes transcriptional suppression through interactions with chromatin-remodeling proteins[1]. LncRNAs, when attached to the chromatin, can regulate the expression of the genes by either adjusting the local chromatin structure at these loci or by directing the regulatory molecules[5]. cis-lncRNAs (cis-acting lncRNAs), through transcriptional interference or via the modification of the chromatin, regulate gene expression in the close genomic vicinity, whereas trans-lncRNAs (i.e., trans-acting lncRNAs) regulate gene expression in distant genomic locations[4]. LncRNAs can also contain specific domains for binding to various effector molecules, acting as scaffolds and assembly units for protein complexes that may have the ability to activate or inhibit gene transcription[6]. A summary of the functions of lncRNAs is shown in Figure 2.
Figure 2. Functions of LncRNAs. RNP: Ribonucleoprotein; lncRNA: long non-coding RNA. Republished with permission from[159].
CELLULAR LOCALIZATION OF LncRNAs
LncRNAs can be present in the cytoplasm, nucleus, or both[13]. They are especially abundant in the nucleus, interacting with the chromatin, where they typically act as epigenetic modulators to control gene transcription[14]. Surprisingly, despite often being more plentiful, nuclear lncRNAs possess less stability than their cytoplasmic counterparts[15]. The hypothesis that the instability of nuclear lncRNAs is controlled by the polyadenylate-binding nuclear protein 1, leading to hyperadenylation and decay via the activity of the enzyme polyA-polymerase, has been put forth as a potential mechanism for the regulation of gene expression. LncRNA levels can thus be regulated via this instability, which allows for flexible regulation in response to stimuli[16,17].
The role of lncRNAs within the cytoplasm, however, would be different from their nuclear counterparts, as cytoplasmic lncRNAs can play a role in the translation of proteins, signal transduction pathways, and regulating gene expression at the post-transcriptional level[15]. For instance, lncRNAs can help in sequestering microRNAs[18], regulating the activity and levels of proteins[19], affecting posttranslational modifications of proteins[20], or influencing mRNA stability[21]. RNA-binding proteins can also be affected by lncRNAs located within the cytoplasm, either through regulating the degradation pathways of proteins via the Ubiquitin pathway or by separating these proteins from their other effector proteins, thus affecting their activity[14].
Interestingly, some studies have investigated the distribution of lncRNAs in other organelles in a cell and tissue-specific expression patterns. The mitochondrial genome has been found to express some genes of lncRNAs, and those genes have been found to be under the regulation of nuclear-encoded proteins[22]. A study performed by Noh et al. found that the lncRNA known as “RMRP” is mainly transcribed from the nuclear genome. However, lncRNA RMRP can subsequently be localized to the mitochondria, where it contributes to integral structure and mitochondrial oxidative phosphorylation[23]. Other studies demonstrated that lncRNAs can also be found within the endoplasmic reticulum, and despite the lack of knowledge about their role and function, these findings represent an interesting fact that needs to be further explored[24,25].
TISSUE AND CELL-SPECIFIC EXPRESSION
Despite usually having lower expression levels compared to mRNAs, lncRNAs tend to display more distinctive expression patterns specific to certain tissues and cells, suggesting that they play crucial roles in cell-specific unique pathways[15]. Nonetheless, there are also housekeeping lncRNAs, which are ubiquitously transcribed across various tissues and cells[26]. According to a study conducted by Derrien et al., while the expression patterns of lncRNAs are more tissue-specific than protein-coding genes, approximately 11% of lncRNAs are identified in every tested tissue, such as TUG1, which is expressed across all tissue types[27,28]. Interestingly, a noteworthy observation reveals a positive correlation between the relative expression of LncRNAs and the breadth of their distribution across tissues. Ubiquitous lncRNAs exhibit high expression levels, while those expressed in specific tissues tend to have comparatively lower expression within the overall lncRNA transcriptome[26]. The following are some of the most widely accepted techniques that are believed to modify the gene expression patterns of lncRNAs.
Histone modifications
One of the most crucial techniques to directly modify differential gene expression depending on the cell type is the chromatin state, which includes histone modification and DNA methylation[29]. The bivalent chromatin domain is a term that defines a state of chromatin methylation with the co-localization of H3K4me3 and H3K27me3 on histones of the same DNA region, either on the same histone molecule or another H3 molecule in the same nucleosome. H3K4me3 is a positive transcription regulator, whereas H3K27me3 is a transcription repressor. This phenomenon is believed to regulate the expression of developmental genes, enabling them to be activated or suppressed in the future, depending on the cell's destiny during differentiation[30]. Several bivalent genes that lose H3K27me3, the transcription repressor, are activated when Embryonic Stem Cells transform into neural progenitor cells, whereas other genes that preserve bivalency or lose H3K4me3, the transcription activator, remain silenced[31]. This behavior determines the genetic expression pattern based on cell and tissue type, and it is possible that this is the reason for the tissue- and cell-specific expression patterns of lncRNAs, which may be derived from bivalent promoter genes[29].
DNA methylation
DNA methylation can be considered a major epigenetic technique to alter the gene expression pattern for any coding or non-coding gene. According to the CpG dinucleotide content of the genome, there are two types of regions: low CpG regions and high CpG regions[32]. Within CpG dinucleotides, DNA methylation mainly occurs at the cytosine, and methylated cytosines often quickly mutate into thymines. Low CpG sites are created when CpG dinucleotides degrade due to mutation after DNA hypermethylation. High CpG and low CpG sites both represent hypomethylation and hypermethylation, respectively. Genes that are located within the low CpG sites may have lower expression levels than genes that are present within the high CpG sites because promoter hypermethylation often suppresses gene expression[33]. Consequently, the lower expression of lncRNAs may be explained by the fact that many lncRNA genes are located within low CpG sites, and what also implicates this fact is the lower perseverance of H3K4me3 around those sites[29].
Transcriptional regulation
At the transcription level, the most diverse and not fully understood mechanism pertains to the regulation of lncRNAs. One example is the shared transcription factors between some lncRNAs and protein-coding genes. For instance, RP1-473L15.2 and ENST00000513542, two lncRNAs, possess motifs corresponding to serum response factor and activating protein-1, respectively[34]. Another example is a novel lncRNA related to DNA damage known as AK019103, which, in its promoter region, exhibits five binding sites for the transcription factor NF-κB[35]. Additionally, core transcription factors such as p53, NF-κB, Sox2, Pou5f1, and Nanog have been found to be sufficient for driving the expression of various lncRNAs across processes ranging from embryonic stem cell pluripotency to cell proliferation[36]. This phenomenon may, in part, reflect the tissue and cell-type specificity inherent in lncRNAs, wherein simultaneous regulation of common genes sharing the same transcription sites occurs in specific tissues and cells where these transcription sites are specifically expressed.
MicroRNAs, a class of ncRNAs smaller in size than lncRNAs and capable of regulating the process of gene expression of protein-coding genes, have also been found to be involved in the transcriptional regulation of lncRNAs[29]. For instance, miRNA-29 has been reported to enhance the expression of the lncRNA MEG3 by inhibiting DNA methyltransferase and preventing methylation of MEG3's promoter[37]. Another interesting example involves miRNA-372, which has been found to regulate the expression of the lncRNA HULC in liver cancer through its interaction with the cAMP Response Element-Binding transcription factor[38]. This indicates that regulatory control over lncRNAs is not exclusive to protein transcription factors; instead, it also involves other factors and mechanisms that are specific to cells and tissues.
Post-transcriptional regulation
Competitive endogenous RNA networks are intricate systems comprising microRNAs, pseudogenes, protein-coding genes, and lncRNAs. These networks feature microRNA response elements (MREs) and engage in crosstalk, mutually influencing the expression and stability of each component interactively[39]. For instance, in human cancer cells, as a post-transcriptional regulatory mechanism, the repression of the lncRNA HOTAIR by miRNA-141 occurs in the presence of the Argonaut 2 (AGO-2) complex[40]. In another study, it was shown that the degradation of the lncRNA HOTAIR involves a collaborative action of miRNA-let7i, HuR, and AGO-2[41]. Moreover, a study led by Han et al. verified that during the development of bladder cancer, the lncRNA MALAT1 experienced downregulation mediated by has-miR-125b[42].
Post-transcriptional regulation of lncRNAs can also involve collaborative mechanisms that enhance the stability of two or more lncRNAs. A notable example is the collaborative action observed between lncRNAs MALAT1 and MENβ, forming a protective triple helix that shields these transcripts from exonuclease damage[43,44].
The stability of lncRNAs can also be influenced by the types and contents of nucleotides they contain. For example, lncRNAs encompass more than 100 distinct modified nucleotides beyond the four core nucleotides found in RNAs. Similar modified nucleotides are present in other short non-coding RNAs such as rRNAs, tRNAs, and small nuclear RNAs and contribute to the stability of these ncRNAs, including lncRNAs[45]. Interestingly, the secondary structures of lncRNAs can serve as substrates for the adenosine deaminase enzyme, catalyzing Adenosine to Inosine (A-to-I) conversion in an RNA editing process. These edited lncRNAs may undergo different processes, including RNA degradation[46].
LncRNAs IN MEDICINE
LncRNAs, with their involvement in various physiological functions and distinct tissue and cell-specific expression, play a pivotal role in the pathology and potential treatment of various diseases [Figure 3]. Here, we briefly highlight a few instances of their involvement as pathological or biological markers (biomarkers) and their potential applications in treatment approaches.
Figure 3. Biomarker Significance and Therapeutic Prospects of LncRNAs in Brain and Spinal Cord Injuries, Prostate Cancer, Breast Cancer, Huntington’s Disease, and Cardiovascular Disorders.
LncRNAs as biomarkers
A biomarker, a biological molecule presents in blood, other bodily fluids, or tissues, serves as an indicator of normal or abnormal processes, conditions, or diseases. Biomarkers are crucial for assessing how effectively the body responds to treatments and the current diagnostic state of any disease[47]. In the specific context of lncRNAs, they can be regarded as biomarkers owing to their tissue-specific genetic characteristics, enabling lncRNAs to elucidate the distinct functions of tissues, making them valuable markers specific to particular tissues and diseases[26,28].
Brain injury
Brain injuries, such as traumatic brain injury (TBI), can result in chronic disabilities and a reduction in survival rate, making them significant health problems[48-50]. TBIs cause acute brain damage in which blood vessels, neurons, and glia stretch or rupture, causing apoptosis and harming the Blood-Brain Barrier. Afterwards, a pro-inflammatory immune response takes place, which causes phagocytosis of dead and injured cells and the secretion of pro-inflammatory cytokines. Chronic inflammation results if this immune response is not resolved in moderate to severe injuries, and its complications can last for weeks to years after injury[48,49,51]. A study conducted by Wang et al. discovered a group of 271 differentially expressed lncRNAs in the rat hippocampus following TBI, and these lncRNAs were associated with several biological functions, including inflammatory responses and apoptosis[52]. Another study on mice discovered 823 abnormally expressed lncRNAs, with 156 downregulated and 667 upregulated genes[53]. In human subjects, Yang et al. investigated the expression of lncRNAs in three different human patients diagnosed with TBI, and the results indicated 56 downregulated and 43 upregulated lncRNAs[54].
Spinal cord injury
Spinal cord injury (SCI) is a devastating disorder of the central nervous system that often leads to the loss of motor and sensory functions, with an incidence rate that ranges from 3.6 to 195.4 cases per million worldwide[55]. The two leading causes of SCI are falls and accidents[56]. SCI, however, can also result from non-traumatic diseases such as inflammatory wounds, poor blood supply, osteoarthritis, and spinal tumors[57]. Paralysis, sensory impairment, neuropathic pain, and other dysfunctions are common among SCI survivors[58]. Earlier studies from Wang et al., using RNA-seq, found seven lncRNAs to be differentially expressed, of which lncSCIR1 was constantly downexpressed at all time points following SCI[59]. Other expression analysis studies revealed 772[60], 277[14], and 458[61] differentially expressed lncRNAs in SCI either at acute SCI or later time points, and those lncRNAs were linked to regulatory functions in immune responses, signaling pathways, and epigenetic modifications[14].
Alzheimer's disease
Alzheimer's disease (AD) is one of the most prevalent neurodegenerative diseases, known for its detrimental effects on cognition and memory[62]. One of the unique neuropathological characteristics of AD is the deposition of the amyloid beta proteins into plaques in the brain parenchyma and cerebral blood vessel walls, and their levels were strongly correlated with cognitive decline in AD[63]. BACE1 (Beta-site Amyloid Precursor Protein (APP) Cleaving Enzyme 1), also known as the β-secretase enzyme, is a type 1 transmembrane aspartic protease enzyme with the highest expression levels found in neurons. It is one of two enzymes, β- and γ-secretase, that acts sequentially on APP (amyloid precursor proteins) for the cleavage and production of amyloid beta proteins[64]. By stabilizing BACE1 mRNA, lncRNA BACE1-AS increases BACE1 protein expression and enhances the formation of amyloid beta proteins[65]. Several other lncRNAs have been found to be dysregulated in AD, such as GDNFOS1[66], 17A[67], NAT-Rad18[68], BC200[69], 51A[70], NDM29[71], and EBF2-AS[72].
Huntington's disease
Huntington's disease (HD) is a rare autosomal dominant neurodegenerative disorder that arises from a repeat expansion of three nucleotides within the Huntingtin gene[73], which results in the aggregation of mutant Huntingtin protein[74]. HD is characterized by motor impairment accompanied by behavioral dysfunction and dementia, in extreme cases, that result from progressive cognitive impairment[75]. It has been found that lncRNA HTT-AS-v1, one of the splice variants of the HTT-AS locus, is markedly downregulated in the brains of HD patients. Moreover, studies have shown that lncRNA HTT-AS-v1 decreases the expression of the HTT gene, which is known as the hallmark of the disease[76]. Another study by Sunwoo et al. showed that about 181 lncRNAs were dysregulated in HD brains, of which 35 were upregulated and 146 were downregulated[77]. In the same study, Sunwoo et al. validated the overexpression of lncRNA NEAT1 and revealed that it is involved in mechanisms of neuroprotection against neuronal damage[77].
Prostate cancer
Prostate cancer, which is the second most common type of carcinoma and the fifth leading cause of cancer mortality among men globally, is primarily composed of adenocarcinomas, a type of cancer that originates from glandular cells. This type of cancer, similar to other types of malignancy, is characterized by having different molecular subtypes, each with its own unique progression pathways[78]. Research has shown that the concentrations of the lncRNA NEAT1 are remarkably higher in tumor tissues and that its levels can predict the overall survival of patients. Additionally, in the context of prostate cancer, it has been discovered that lncRNA NEAT1 has the ability to change the epigenetic environment and promote the active transcription of genes[79]. Another recent study by Yuan et al. unveiled the role of the overexpressed lncRNA SSTR5-AS1 in prostate cancer as a prognostic marker that can alter the level of proteins involved in the proliferation and metastasis of prostate cancer cells[80].
Breast cancer
According to the World Health Organization, breast cancer (BC) is the most common type of cancer that affects women worldwide[81], with more than 2.3 million cases and 685,000 fatalities worldwide[82]. A study by Sang et al. has revealed that when the lncRNA PANDAR is expressed at higher levels in breast cancer cells, it helps the cells multiply by controlling the transition from G1 to S phase[83]. Another lncRNA that has shown a role in the determination of the prognosis for breast cancer is lncRNA linc00511. This lncRNA has been investigated for its role in promoting tumor growth, accelerating the transition from the G1 to the S phase of breast cancer cells, and inhibiting apoptosis[84].
Cardiovascular disease
Chronic heart failure (CHF), broadly categorized as either ischaemic or non-ischaemic cardiomyopathy, is a multifaceted clinical syndrome arising from diverse structural or functional impediments in ventricular filling or blood ejection. It stands as a prominent cause of mortality, particularly affecting the aging population, with an estimated lifetime risk of developing CHF ranging from 20% to 33%. Despite advancements in therapies, approximately half of CHF-diagnosed patients face a grim prognosis, with a significant number succumbing within five years[85]. Recent studies have brought attention to numerous aberrantly expressed lncRNAs in CHF, shedding light on their potential roles and significant functions in both the pathology and potential alleviation of the disease[85-88]. For instance, in a study by Li et al. focusing on dilated cardiomyopathy-induced CHF, lncRNA profiling identified 20 upregulated and 20 downregulated lncRNAs. Among them, the upregulated lncRNA RP11-544D21.2 and the downregulated lncRNA XLOC_014288 were correctly validated and functionally characterized[85]. In another investigation by Zhang et al., the lncRNA cytb was discovered to be downregulated in the heart and plasma of patients with heart failure[86].
LncRNAs as potential therapeutic approaches
Brain injury
As previously mentioned, lncRNAs can be differentially expressed in brains following TBI, affecting many biological processes that follow brain injuries. Zhong et al. investigated the differentially expressed lncRNA NEAT1 through knockdown experiments in mice and found that NEAT1 was found to decrease the number of cells undergoing apoptosis and decrease the intensity of inflammation[89]. In a study by
Spinal cord injury
As a promising therapeutic strategy, neural stem cells have been proposed as a potential solution[91]. It has been shown that the lncRNA UCA1 controls the differentiation and proliferation of neural stem cells, and therefore, it can have a huge impact on the treatment of SCI[92]. Another interesting study has shown that hydrogen sulfide, a potential treatment for SCI, has an impact on the upregulation of lncRNA CasC7, which results in decreasing the apoptosis of neuronal cells and improving SCI[93].
Alzheimer's disease
Gene silencing with siRNAs is a method of knocking down genes that results in gene expression regulation[94]. The introduction of siRNAs, which target the lncRNA BACE1-AS, into the transgenic AD mouse brains reduced the expression of both BACE1-AS and BACE1[95]. This approach poses the question of whether the same can be applied to many other dysregulated lncRNAs that are involved in many pathophysiological pathways.
Huntington's disease
One of the repressed genes in HD is the protein Brain-Derived Neurotrophic Factor (BDNF)[96]. BDNF enhances the survival of the neurons and their cytoskeleton rearrangement, providing neuroprotection during inflammation[97]. It has been hypothesized that the loss of BDNF may explain the loss of neurons where mutant Huntingtin protein is ubiquitously expressed[98,99]. The presence of lncRNA BDNF-AS has been found to decrease the expression and function of the BDNF protein via chromatin modification at the BDNF gene location and thus suppress the expression of the BDNF transcript[100]. Therefore, studying the expression of lncRNA BDNF-AS in HD may be a promising point that can be used for the design of a therapeutic approach for HD.
Prostate cancer
The potential therapeutic effects of lncRNAs in prostate cancer have been growing recently. A recent study has shown that the lncRNA DNMBP-AS1, which is upregulated in prostate cancer, is capable of inducing the cellular proliferation and metastasis of prostate cancer cells via sponging of miR-6766-3p. Knocking down this lncRNA has led to the inhibition of cancer cell proliferation and metastasis[101]. Another earlier study investigated the effect of the overexpression of already downregulated lncRNAs; this study has shown that the upregulation of the expression of the lncRNA MIR22HG in prostate cancer cells, which resulted in the sequestration of miR-9-3p, led to the inhibition of apoptosis and cancer cell proliferation[102].
Breast cancer
It has been shown that the downexpression of lncRNA PANDAR, using a siRNA-based technique, led to a reduction in the cellular growth of in vitro breast cancer cells and increased the number of cells within the G1 phase[83]. Another interesting study by Ren et al. discovered that inhibition of TGF-β1 along with other regulatory mediators resulted in the downregulation of the lncRNA HOTAIR and its action on inducing breast cancer metastasis[103].
Cardiovascular disease
The potential therapeutic impact of lncRNAs in cardiovascular disease has been substantiated in numerous research studies. For instance, in a study conducted by Li et al., a loss-of-function silencing assay was performed on the dysregulated lncRNA RP11-544D21.2, which was identified to be upregulated in CHF model cells. The findings revealed that the silencing of RP11-544D21.2 had a significant impact on various downstream pathways, including the arrhythmogenic right ventricular cardiomyopathy pathway, hypertrophic cardiomyopathy, antigen processing and presenting pathway, cell adhesion pathway-related genes, cell cycle, focal adhesion, and Extracellular Matrix (ECM)-receptor interaction[85]. Remarkably, lncRNA ZNF593-AS was also discovered to alleviate contractile dysfunction not only in dilated cardiomyopathy but also in diabetic cardiomyopathy[87,88]. These findings provide additional evidence for the potential therapeutic benefits of lncRNAs in the context of cardiovascular disease.
RNAs IN DRUG DEVELOPMENT
Recently, researchers have begun to target RNAs as potential drug targets because they make up a large portion of the genome and offer greater specificity than proteins. LncRNAs, in particular, may be a more precise target than epigenetic complexes. RNA-based therapies also have the advantage of being highly specific and stable[14]. RNA-based drugs fall into the category of oligonucleotide-based drugs, which can be classified into two main categories based on their chemical structure: double-stranded RNA drugs, such as siRNA, and single-stranded DNA drugs[104]. RNA molecules, such as antisense oligonucleotides, siRNAs, microRNAs, and lncRNAs, may target mRNAs and ncRNAs with specificity through Watson-Crick base-pairing. Antisense oligonucleotides, being the most common, have two different mechanisms of action: RNA interference (RNAi) and RNase-H-mediated degradation[105].
Antisense oligonucleotides
Antisense oligonucleotides (ASOs) are unpaired DNA strands that have complete complementarity for a single target mRNA molecule[106]. The targeting of mRNAs or microRNAs through the use of ASOs results in the degradation of the target through the activity of the RNase H endonuclease enzyme. This process leaves the ASOs intact, allowing them to act as catalysts in the reaction and affect multiple target molecules[107]. These ASO-based drugs work by influencing the production or function of disease-causing proteins through changes in pre-mRNA splicing, mRNA degradation, regulation of protein translation, or direct interactions with proteins[108]. ASOs need to be administered repeatedly because the oligonucleotides are broken down by the cellular nucleases over time[109].
MicroRNAs
MicroRNAs are a class of ncRNAs that are small in size and capable of regulating the process of gene expression of protein-coding genes post-transcription. They exist naturally, with over 2,300 different microRNAs present in human cells, whose expression patterns differ depending on the tissue and time[110]. They form an imperfect RNA-hairpin structure due to regions of complementarity. When processed by the RNA interference machinery, they create a 19-25 nucleotide RNA duplex with mismatching regions. microRNAs normally bind to the 3' untranslated region (3' UTR) of mRNA and prevent protein translation. Unlike siRNAs, which require virtually full sequence complementarity to cause cleavage of the target RNA, microRNAs only need a small number of complementary nucleotides to the target mRNA in order to inhibit its translation; therefore, microRNAs can have several targets throughout the cell[109]. microRNAs can be used as potential therapeutic approaches, as follows.
MicroRNA mimics
MicroRNA mimics are artificial RNAs that are built upon naturally occurring microRNAs. The so-called “passenger” strand of the mimic, which is not loaded into the RNA-induced silencing complex (RISC), carries some mismatches; this is to prevent it from being loaded into RISC and to prevent its potential action as an anti-microRNA[111].
Anti-microRNAs/AntagomiRs
Anti-microRNAs are oligonucleotides with sequences that are either entirely or partially complementary to naturally occurring microRNAs. Consequently, this leads to blocking the interaction with the target genes when they interact with the endogenous microRNA[106].
MicroRNA sponges
MicroRNA sponges are synthetic transcripts that contain multiple binding sites for microRNAs, which can trap and sequester them[112]. They can be designed to target one or many different microRNAs[113,114]. A study by Jung et al. designed microRNA sponges that can sequester miR-155, miR-21, and miR-221/miR-222 within tumor-causing cells[115]. Another study by Das et al. investigated the role of microRNA sponges within myocardial infarction by designing anti-microRNA that can sequester a family of miR-181 microRNAs, which are miR-181a, miR-181b, and miR-181c[116].
MicroRNA masks
MicroRNA masks are oligonucleotides that are designed to conceal the microRNA binding regions of a target gene. This prevents the microRNAs from carrying out their regulatory functions on that gene[117]. TYRP1 mRNA acts to sequester miR-16, inhibiting their tumor-suppressing functions within melanoma cells. One study by Gilot et al. demonstrated the effects of blocking the microRNA binding site of TYRP1 mRNA, which caused the recovery of the cancer-suppressing function of miR-16 in melanoma tumors[118].
Small interfering RNAs
Small interfering RNAs (siRNAs) are short double-stranded molecules created by an enzyme called Dicer. This enzyme cuts a long double-stranded RNA into smaller pieces known as siRNA, which are between 21 and 23 base pairs long and have a low molecular weight of around 14 kDa[119]. They make use of the natural endogenous microRNA pathway[106]. A molecule of siRNA consists of both a sense and an antisense strand that form complexes by binding to the RISC. The sense strand separates from the antisense strand with the help of AGO-2. The antisense strand then guides the RISC to find a complementary mRNA, where it is then cut by the AGO-2-RISC-siRNA complex, effectively stopping the translation process. They work by initiating the process of breaking down mRNA, silencing specific genes, and reducing the levels of target-specific proteins[119]. Unlike antisense techniques, RNAi uses a catalytic mechanism for its action, where siRNA-loaded RISC can dissociate and bind to other mRNA molecules after the target mRNA is cleaved. This is why it is effective at very low concentrations of siRNA in the picomolar range, which can efficiently knock down genes, and it has been found that intracellular amounts of less than 2,000 siRNAs per cell are enough to achieve this effect. It is also important to note that the siRNA mechanism is different from that of microRNAs, which bind to their targets with only partial complementarity, whereas siRNA action requires 100% complementarity[120].
Short hairpin RNAs
Short hairpin RNAs are single-stranded RNAs made up of two sets of complementary sequences on the same strand, known as “sense” and “antisense” sequences, that are 19-22 nucleotides in length. These sequences contain a short loop consisting of 7 to 9 nucleotides that are not bonded, which enables them to form a hairpin shape[109]. The main function of short hairpin RNAs is to produce double-stranded siRNAs that can be further used for the degradation of target sequences, as previously explained. This can be accomplished via the introduction of short hairpin RNAs, which are usually expressed using either RNA polymerase II or III enzymes, the same as pri-microRNAs, producing a structure that is similar to pre-microRNA, followed by processing using Dicer and the formation of the mature siRNAs that will exert their biological effect[110]. One limitation of using siRNA for RNA interference is that after siRNA is delivered to target cells, the effect of knocking down a specific gene gradually decreases as the oligonucleotides are decreased in concentration by either cellular division or being broken down by nucleases. However, when delivered on a plasmid vector, shRNA sequences can provide prolonged RNAi over a longer period of time because double-stranded DNA is more resistant to cellular degradation, in addition to their stable positioning in the nucleus as episomes, providing a continuous source of the therapeutic RNAi sequence. Another benefit of using short hairpin RNAs in therapeutics is the ability to selectively lower gene target expression only in cells relevant to the disease by controlling the expression of the shRNA vectors with promoters that can only express the relative genes in specific target cells[109].
Long non-coding RNAs
There are two approaches to lncRNA therapeutics research: the first is to use synthetic oligonucleotides that resemble lncRNA in their structure and their binding properties as well, acting as decoys and interfering with their function, while the second is to target lncRNAs directly[121]. Despite the greater functional ability of lncRNAs, their aberrant roles in pathophysiological processes, and their specific tissue and cellular expression, there has not been, to our knowledge, any role for the introduction of lncRNA-based therapeutics in any preclinical or clinical studies. However, targeting lncRNAs has been the case for some RNA-based studies.
RNA interference using siRNAs, according to a study by Modarresi et al., reported that the inhibition of the lncRNA known as BDNF-AS, a lncRNA antisense transcript that controls BDNF mRNA and protein expression, caused the chromatin status of the BDNF gene to be altered, increased BDNF protein levels, and promoted neuronal differentiation and proliferation both in vitro and in vivo[122]. Recent clinical trials are also looking into the role of antisense oligonucleotides in suppressing the lncRNA UBE3A-ATS, which regulates the expression of Ubiquitin ligase E3A in Angelman syndrome[123]. These are just two of many studies that have targeted lncRNAs for therapeutic treatment, and we are only scratching the surface of our understanding and knowledge of the potential benefits of lncRNA-based therapeutics.
RNA THERAPEUTICS: LAB TO MARKET
The translation of the therapeutic potential of RNAs from preclinical studies into actual RNA therapeutics is a multifaceted process that involves numerous challenges and considerations. Here, we provide a few examples of RNA therapeutics and delve into their potential applications and impacts.
Fomivirsen
Fomivirsen is a synthetic antisense oligonucleotide that gained prominence as an innovative therapeutic agent in the field of ophthalmology. It is formed of a 21-mer phosphorothioate oligonucleotide sequence designed to target the immediate-early 2 gene of Cytomegalovirus. Administered through intraocular injections, this groundbreaking antiviral medication demonstrated significant efficacy in slowing the progression of Cytomegalovirus retinitis, particularly in individuals with compromised immune systems, such as those with HIV/AIDS. Fomivirsen's unique mechanism of action paved the way for a new era in nucleic acid-based therapies, showcasing the potential of antisense technology in treating challenging viral infections[124].
Mipomersen
Mipomersen is a novel therapeutic antisense oligonucleotide agent that has shown promise in addressing Familial Hypercholesterolemia, a genetic disorder characterized by elevated levels of low-density lipoprotein cholesterol in the blood. Familial Hypercholesterolemia is often resistant to traditional lipid-lowering treatments, making the development of alternative approaches crucial. Mipomersen, a 20-mer phosphorothioate oligonucleotide, functions by inhibiting the production of Apolipoprotein B-100 mRNA, a key component of low-density lipoprotein particles. Apolipoprotein B-100 is responsible for the structural integrity of low-density lipoprotein and is directly involved in cholesterol transport. By targeting Apolipoprotein B-100 mRNA, Mipomersen helps reduce low-density lipoprotein levels, offering a potential solution for individuals with Familial Hypercholesterolemia who face an increased risk of premature cardiovascular disease[125].
Givosiran
Givosiran represents a groundbreaking siRNA therapeutic approach in the treatment of Acute Hepatic Porphyria, which is a rare genetic disorder characterized by the impaired synthesis of heme, a crucial component of hemoglobin. Givosiran, a 21-bp double-stranded siRNA, targets the root cause of the disease by inhibiting Aminolevulinate Synthase 1 (ALAS1) mRNA, the enzyme responsible for the initial step in heme synthesis. By reducing ALAS1 activity, Givosiran helps decrease the accumulation of porphyrin precursors that trigger acute attacks in patients. This RNA interference technology represents a significant advancement in managing the symptoms of the disease, providing a novel and targeted solution to improve the quality of life for individuals affected by this challenging condition[126].
Miravirsen
Miravirsen represents a groundbreaking advancement in the treatment of Hepatitis C (HCV), a viral infection affecting the liver. Developed as an antimiR against miR-122, Miravirsen specifically targets and inhibits miR-122, a host cell microRNA essential for the replication of HCV. MiR-122 plays a crucial role in the viral life cycle by promoting HCV RNA stability and facilitating its translation. Miravirsen disrupts this process by binding to miR-122, preventing its interaction with the viral RNA. This innovative approach effectively suppresses HCV replication, offering a promising therapeutic strategy. Clinical trials have shown encouraging results, establishing Miravirsen as a potential game-changer in the fight against Hepatitis C, with the prospect of improving patient outcomes and reducing the global burden of this infectious disease[127].
Remlarsen
Remlarsen is a novel microRNA mimic therapeutic agent that has shown promising results in the treatment of cutaneous and pulmonary fibrosis. Fibrosis is a condition characterized by the excessive formation of fibrous connective tissue, leading to scarring and impaired organ function. Cutaneous fibrosis affects the skin, while pulmonary fibrosis affects the lungs. One key player in the regulation of fibrosis is miR-29b, a microRNA that acts as a molecular brake on fibrotic processes by inhibiting the expression of pro-fibrotic genes. Research suggests that Remlarsen works by mimicking the structure of miR-29b, thereby mitigating fibrosis in both the skin and lungs. This innovative approach offers hope for individuals suffering from these debilitating conditions, providing a potential breakthrough in the field of fibrotic disease therapeutics[128].
RG-125
RG-125 is a promising therapeutic agent in the context of Non-Alcoholic Fatty Liver Disease, a prevalent and potentially serious liver condition associated with excessive fat accumulation in the liver. RG-125 operates by targeting miR-103/107, microRNAs implicated in the regulation of lipid metabolism and the progression of the disease. These microRNAs play a crucial role in the control of lipid homeostasis, and their dysregulation has been linked to the development of fatty liver disease. RG-125, by modulating
MRX34
MRX34, a miR-34a mimic, represented an innovative approach in the field of RNA therapeutics by aiming to supplement the tumor-suppressive functions of the microRNA known as miR-34a. MiR-34 plays a pivotal role in regulating gene expression, particularly in the context of inhibiting cancer cell proliferation and promoting apoptosis. The initial excitement surrounding MRX34 stemmed from its potential to mimic the actions of miR-34a, thereby harnessing the natural mechanisms that control cellular processes and suppress cancer development. However, the progress of clinical studies with MRX34 was abruptly halted due to unforeseen severe immune-related adverse events observed in multiple patients. This unexpected outcome raised concerns about the safety profile of the miR-34a mimic and underscored the challenges and complexities associated with the development of RNA-based therapeutics in the quest for innovative cancer treatments[130].
A summary of different types of RNA therapeutics and their related commercialized and in-development drugs are shown in Figure 4 and Table 1, respectively.
Figure 4. RNA-based therapeutics. This illustration shows the different types of RNA-based therapeutics. Antisense oligonucleotides can act by degrading paired mRNA through the activity of RNase H endonuclease or by directly binding to proteins through their 3D structure, acting as aptamers. siRNAs can be introduced directly or produced from short hairpin RNAs through the action of the enzyme Dicer. siRNAs then bind with complete affinity to mRNAs and mediate their degradation through RISC cleavage. MicroRNA mimics, on the other hand, bind with partial affinity to mRNAs and follow the same pathway. Other types of microRNAs interfere with this type of binding by either directly binding with microRNAs through oligonucleotides, using sponges to sequester multiple microRNAs at once, or indirectly by blocking the binding site on mRNA targets. lncRNAs can be used to stimulate agonistic functions through mimics or targeted for antagonistic functions. ASOs: Antisense oligonucleotides; LncRNAs: long non-coding RNAs; miRNAs: microRNAs; shRNA: short hairpin RNA; siRNAs: small interfering RNAs; RISC: RNA-induced silencing complex; Dicer: RNAase III enzyme.
Examples of RNA-based therapeutics in medicine
| Type of drug | Drug name | FDA approval | Clinical studies | Active moiety | Target | Disease | Reference |
| Antisense oligonucleotides | Fomivirsen | Yes | Completed | 21-mer phosphorothioate oligonucleotide | IE2 mRNA | Cytomegalovirus | [124] |
| Pegaptanib | Yes | Completed | 28-mer RNA aptamer | VEGF165 protein | Age-related macular degeneration | [131] | |
| Nusinersen | Yes | Completed | 18-mer 2′-O-methoxyethyl aptamer | ISS-N1 protein | Spinal muscular atrophy | [132] | |
| Mipomersen | Yes | Completed | 20-mer phosphorothioate oligonucleotide with 2′-O-methoxyethyl modification | ApoB-100 mRNA | Familial hypercholesterolemia | [125] | |
| Small interfering RNAs | Patisiran | Yes | Completed | 21-bp double-stranded siRNA encapsulated in a lipid nanoparticle | Transthyretin mRNA | Hereditary transthyretin amyloidosis | [133] |
| Givosiran | Yes | Completed | GalNAc-conjugated 21-bp double-stranded siRNA | ALAS1 mRNA | Acute hepatic porphyria | [126] | |
| Lumasiran | Yes | Completed | GalNAc-conjugated 21-bp double-stranded siRNA | HAO1 mRNA | Primary hyperoxaluria type 1 | [134] | |
| Short hairpin RNAs | N/A | No | Not yet | Single-stranded shRNA on a scAAV construct | SOD1 mRNA | Amyotrophic lateral sclerosis | [135] |
| MicroRNAs | MRX34 | No | Halted | miRNA-34a mimic in liposomal nanoparticles | mRNA targets of miR-34a | A variety of malignant or carcinogenic conditions, including hepatocellular carcinoma, ovarian cancer, colon cancer, non-small cell lung carcinoma, and cervical cancer | [130] |
| Miravirsen | No | Phase II | AntimiR against | miR-122 | Hepatitis C | [127] | |
| Remlarsen | No | Phase II | miR-29b mimic | mRNA targets of miR-29b | Cutaneous and pulmonary fibrosis | [128] | |
| RG-125 | No | Phase I/II | AntimiR against | miR-103/107 | Non-alcoholic fatty liver disease | [129] |
CHALLENGES AND LIMITATIONS OF LncRNA THERAPEUTIC DEVELOPMENT
As of now, no lncRNA-based therapeutics have entered clinical trials in any phase. Despite this, the promising potential of lncRNAs as a therapeutic approach suggests that their entry into clinical trials is on the horizon[106,136]. Nevertheless, the development of lncRNA-based therapeutics must navigate challenges and limitations to maximize therapeutic efficacy and ensure the successful translation of these innovative approaches into clinical applications.
Cross-species sequence variation
The sequence variation in lncRNA sequences that is present between humans and animal models used for experimentation can make it difficult to create effective therapeutics. Drugs found through testing on human cells may not work when tested on rodents or other disease models, as lncRNAs may have different functions in humans compared to other animals. To overcome this, animal models might have to be altered to express the same lncRNA present in humans. However, this would be a difficult task as it would require a thorough understanding of the lncRNA and its target gene interactions[137].
Optimum silencing strategy
Silencing genes can be used to study the function of a lncRNA through the use of antisense oligonucleotides or double-stranded RNAs. However, silencing lncRNAs is more complex than silencing mRNA because the subcellular localization of lncRNAs varies and different silencing strategies have varying effectiveness. Antisense oligonucleotides are more effective in targeting RNAs located in the cell nucleus, while double-stranded RNAs are better for RNAs in the cytoplasm. This suggests that if a target RNA is expected to perform a role in the nucleus, antisense oligonucleotides would be the best silencing method, while double-stranded RNAs would be better for targets thought to function in the cytoplasm[138,139].
Structural and molecular diversity
LncRNAs can have several domains that are involved in different interactions; blocking these interactions could have therapeutic value. Understanding the pertinent RNA motifs and their structural complexity is necessary for finding ligands that can bind lncRNAs with high specificity and affinity. Currently, this level of knowledge is only attainable for a small number of lncRNAs. The progress in this field will increase as more information about lncRNAs' structural and molecular features becomes available[121].
Immunogenicity and tolerability
As a defense mechanism against viruses, our immune system identifies both single-stranded and double-stranded RNA through various extracellular and intracellular pathogen-associated molecular pattern receptors, including Toll-like receptors, which can trigger diverse immune responses[140]. Additionally, research has demonstrated that single-stranded RNA is more likely to induce immune stimulation compared to double-stranded RNA. Consequently, all currently used or under-development siRNAs in clinical applications are designed as double-stranded structures[106,141].
The primary pathway for recognizing RNA therapeutics involves Toll-like receptor signaling, mediated through myeloid differentiation factor-88. This activation triggers various pathways, leading to the activation of NF-κB and subsequent production of pro-inflammatory cytokines (such as IL-6, IL-8, IL-12, and TNF). Alternatively, it can induce a type I interferon response, culminating in the activation of diverse downstream immune responses[142]. A notable example is the miR-34 mimic MRX34, which resulted in substantial adverse reactions in five patients during a multicenter phase I clinical trial for individuals with advanced malignancies. These reactions included a case of cytokine release syndrome[143,144].
Specificity and off-target effects
The efficacy of an RNA therapeutic is determined by the precision of its on-target specificity and the absence of off-target and unintended on-target effects[106]. For instance, Oblimersen, a first-generation 18 nucleotide ASO targeting BCL2 mRNA, underwent testing in several clinical trials but exhibited limited effectiveness. An off-target effect was observed as Oblimersen induced apoptosis in BCL-2-negative cell lines[145]. Interestingly, BCL2-targeting Oblimersen demonstrated various off-target effects, including the upregulation of stress-response genes, downregulation of proliferation-associated genes, and reduced levels of several apoptosis- and glycolysis-related proteins[146,147]. Another instance of an off-target effect is the termination of the anti-miR-122 therapeutic RG-101 due to elevated levels of bilirubin in the blood[148].
Delivery methods
The efficient delivery of RNA therapeutics, ensuring they reach the intended organ and cell type and successfully traverse the cell membrane to exert their intracellular functions, remains a formidable challenge in the field. Various strategies address the formulation of RNA therapeutics and their delivery mechanisms. Nucleotide modifications play a role in enhancing stability by evading nucleases and reducing interactions with proteins. Delivery mechanisms encompass lipid nanoparticles, polymers like polyethylene imine, polylactic-co-glycolic acid, poly-amidoamine, and chitosan. Additionally, conjugation to active molecules, such as N-acetylgalactosamine, is employed to enhance delivery efficiency[106].
Toxicity
Toxicity is another crucial consideration in the administration of RNA therapeutics. For instance, it has been demonstrated that off-target effects are more sensitive to small concentrations of siRNAs than on-target silencing[149]. Many RNA therapeutics leverage endogenous RNA interference machinery, and excessive dosing can lead to system saturation, impeding the function of endogenous microRNAs. The initial report of this phenomenon indicated that robust shRNA overexpression in hepatocytes resulted in a global downregulation of microRNAs, leading to liver toxicity and mortality in mice[150]. A comprehensive screening study confirmed that the global upregulation of microRNA target genes is a general occurrence triggered by the introduction of exogenous small RNAs[151].
Undesired on-target effects can contribute to the toxicity of RNA therapeutics. For example, in the case of MRX34, it was observed that the therapeutic was taken up by white blood cells[143]. Since miR-34a plays a significant role in T cells and macrophages, incubation with a miR-34a mimic led to changes in chemokine profiles in macrophages and T cells[152]. This is believed to be a factor in the low response rate and adverse immune effects observed in the clinical trial of MRX34[106]. Similarly, the increased chemotherapy-induced neuropathy in patients receiving the AEG35156 ASO may be attributed to the downregulation of the XIAP gene not only in tumor cells but also in oligodendrocytes, glial cells, or neuronal cells, leading to these undesirable effects[153].
CONCLUDING REMARKS
LncRNAs are a class of RNA transcripts that are long (i.e., more than 200 nucleotides) and not translated into proteins[6]. LncRNAs vary considerably in function and play critical roles in many pathophysiological processes, such as RNA splicing and X-chromosome inactivation[1,10]. LncRNAs often exhibit low expression levels and are typically associated with specific tissues or cell types[7]. They have been growing as new contributors to cancer in both oncogenic and tumor suppressive pathways[8]. Being involved in various physiological functions, lncRNAs can be involved in pathology and the potential treatment of different diseases.
LncRNAs have shown their involvement as either pathological markers and/or therapeutic potential targets in diseases such as brain injury[52-54], spinal cord injury[14,59-61], Alzheimer’s disease[65-67], Huntington’s disease[76,77], prostate cancer[79,80], breast cancer[83,84], and many other diseases[154-158]. Recently, researchers have begun to target RNAs as potential drug targets or formulate RNA-based drugs for which they offer greater specificity and stability than proteins[14]. Different RNA-based drugs include antisense oligonucleotides[106], microRNA mimics[111], anti-microRNAs[106], microRNA sponges[112], microRNA masks[117], small interfering RNAs[119], short hairpin RNAs[109], and long non-coding RNAs[121]. Different examples of RNA-based drugs, either in preclinical or clinical trials, can be shown in Table 1.
However, despite the greater functional ability of lncRNAs, there has not been any attempt to formulate lncRNA-based drugs, and they have not entered clinical trials. Nonetheless, lncRNAs have been used as a therapeutic target in many studies[122,123]. The complications that can face the development of lncRNA-based drugs are the sequence variation in lncRNA sequences that is present between humans and animal models used for experimentation[137], the variation of the subcellular localization of lncRNAs, which requires different silencing strategies with varying effectiveness[138,139], the limited knowledge about the structural complexity of lncRNAs[121], and the immunogenicity of some RNAs, which may elicit pro-inflammatory responses and diminish therapeutic efficacy[143,144]. Additionally, the presence of off-target effects poses a risk of causing adverse effects in patients[106]. Toxicity-related concerns arise from undesirable off-target effects or on-target non-specific effects[149,152]. Lastly, the choice of an appropriate delivery method is a critical consideration in developing lncRNA-based therapeutics[106].
Finally, we conclude that lncRNA research is still in progress and that there is a huge potential in the field of lncRNA therapeutics, either as drug targets or lncRNA-based drugs, which can alter the way we understand and treat both common and rare diseases.
DECLARATIONS
Authors’ contributions
Carried out the review’s conceptualization and design: Kamal A, El-Nahrery EM, Swellam M, Shalaby NM, Darwish MK
Drafted the manuscript: Kamal A, El-Nahrery EM
Reviewed the manuscript for important intellectual content: Swellam M
The final manuscript that will be published has been read and approved by all authors.
Availability of data and materials
Not applicable.
Financial support and sponsorship
None.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2024.
REFERENCES
1. Malik B, Feng FY. Long noncoding RNAs in prostate cancer: overview and clinical implications. Asian J Androl 2016;18:568-74.
2. Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers 2016;2016:9085195.
3. Jiang X, Yan Y, Hu M, et al. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg 2016;124:129-36.
6. Xi J, Sun Q, Ma L, Kang J. Long non-coding RNAs in glioma progression. Cancer Lett 2018;419:203-9.
7. Yang M, Zhai Z, Guo S, Li X, Zhu Y, Wang Y. Long non-coding RNA FLJ33360 participates in ovarian cancer progression by sponging miR-30b-3p. Onco Targets Ther 2019;12:4469-80.
8. Shao M, Liu W, Wang Y. Differentially expressed LncRNAs as potential prognostic biomarkers for glioblastoma. Cancer Genet 2018;226-7:23-9.
9. Kiang KMY, Leung GKK. Clinical significance of CRNDE transcript variants in glioblastoma multiforme. Noncoding RNA Res 2017;2:119-21.
10. Ma Y, Luo T, Dong D, Wu X, Wang Y. Characterization of long non-coding RNAs to reveal potential prognostic biomarkers in hepatocellular carcinoma. Gene 2018;663:148-56.
11. Zeng T, Li L, Zhou Y, Gao L. Exploring long noncoding RNAs in glioblastoma: regulatory mechanisms and clinical potentials. Int J Genomics 2018;2018:2895958.
12. Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res 2018;3:108-17.
13. Cabili MN, Dunagin MC, McClanahan PD, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 2015;16:20.
14. Duran R, Wei H, Kim DH, Wu JQ. Invited review: long non-coding RNAs: important regulators in the development, function and disorders of the central nervous system. Neuropathol Appl Neurobiol 2019;45:538-56.
15. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol 2021;220:e202009045.
16. Clark MB, Johnston RL, Inostroza-Ponta M, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res 2012;22:885-98.
17. Bresson SM, Hunter OV, Hunter AC, Conrad NK. Canonical Poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet 2015;11:e1005610.
18. Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun 2016;7:10982.
19. Grelet S, Link LA, Howley B, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol 2017;19:1105-15.
20. Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol 2016;18:213-24.
21. Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 2011;470:284-8.
22. Rackham O, Shearwood AM, Mercer TR, Davies SM, Mattick JS, Filipovska A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011;17:2085-93.
23. Noh JH, Kim KM, Abdelmohsen K, et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev 2016;30:1224-39.
24. Kaewsapsak P, Shechner DM, Mallard W, Rinn JL, Ting AY. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. Elife 2017;6:e29224.
25. Fazal FM, Han S, Parker KR, et al. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 2019;178:473-90.e26.
26. Jiang C, Li Y, Zhao Z, et al. Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 2016;7:7120-33.
27. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89.
28. Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 2014;24:616-28.
30. Macrae TA, Fothergill-Robinson J, Ramalho-Santos M. Regulation, functions and transmission of bivalent chromatin during mammalian development. Nat Rev Mol Cell Biol 2023;24:6-26.
31. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006;125:315-26.
32. Beck D, Ben Maamar M, Skinner MK. Genome-wide CpG density and DNA methylation analysis method (MeDIP, RRBS, and WGBS) comparisons. Epigenetics 2022;17:518-30.
33. Elango N, Yi SV. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol 2008;25:1602-8.
34. Song G, Shen Y, Zhu J, et al. Integrated analysis of dysregulated lncRNA expression in fetal cardiac tissues with ventricular septal defect. PLoS One 2013;8:e77492.
35. Wan G, Hu X, Liu Y, et al. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J 2013;32:2833-47.
36. Dinger ME, Amaral PP, Mercer TR, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008;18:1433-45.
37. Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011;30:4750-6.
38. Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83.
39. Shi X, Sun M, Wu Y, et al. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumour Biol 2015;36:503-13.
40. Chiyomaru T, Fukuhara S, Saini S, et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem 2014;289:12550-65.
41. Yoon JH, Abdelmohsen K, Kim J, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 2013;4:2939.
42. Han Y, Liu Y, Zhang H, et al. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long noncoding RNA MALAT1. FEBS Lett 2013;587:3875-82.
43. Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 2012;26:2392-407.
44. Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci USA 2012;109:19202-7.
45. Cantara WA, Crain PF, Rozenski J, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res 2011;39:D195-201.
46. Yang Y, Zhou X, Jin Y. ADAR-mediated RNA editing in non-coding RNA sequences. Sci China Life Sci 2013;56:944-52.
48. Yang Y, Ye Y, Su X, He J, Bai W, He X. MSCs-derived exosomes and neuroinflammation, neurogenesis and therapy of traumatic brain injury. Front Cell Neurosci 2017;11:55.
49. Kim DK, Nishida H, An SY, Shetty AK, Bartosh TJ, Prockop DJ. Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc Natl Acad Sci USA 2016;113:170-5.
50. Shi Y, Shi H, Nomi A, Lei-Lei Z, Zhang B, Qian H. Mesenchymal stem cell-derived extracellular vesicles: a new impetus of promoting angiogenesis in tissue regeneration. Cytotherapy 2019;21:497-508.
51. Roura S, Monguió-Tortajada M, Munizaga-Larroudé M, et al. Potential of extracellular vesicle-associated TSG-6 from adipose mesenchymal stromal cells in traumatic brain injury. Int J Mol Sci 2020;21:6761.
52. Wang CF, Zhao CC, Weng WJ, et al. Alteration in long non-coding RNA expression after traumatic brain injury in rats. J Neurotrauma 2017;34:2100-8.
53. Zhong J, Jiang L, Cheng C, et al. Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res 2016;1646:589-600.
54. Yang LX, Yang LK, Zhu J, Chen JH, Wang YH, Xiong K. Expression signatures of long non-coding RNA and mRNA in human traumatic brain injury. Neural Regen Res 2019;14:632-41.
55. Shen H, Fan C, You Z, Xiao Z, Zhao Y, Dai J. Advances in biomaterial-based spinal cord injury repair. Adv Funct Mater 2022;32:2110628.
56. Badhiwala JH, Wilson JR, Fehlings MG. Global burden of traumatic brain and spinal cord injury. Lancet Neurol 2019;18:24-5.
57. Hatch BB, Wood-Wentz CM, Therneau TM, Walker MG, Payne JM, Reeves RK. Factors predictive of survival and estimated years of life lost in the decade following nontraumatic and traumatic spinal cord injury. Spinal Cord 2017;55:540-4.
58. Ramer LM, Ramer MS, Bradbury EJ. Restoring function after spinal cord injury: towards clinical translation of experimental strategies. Lancet Neurol 2014;13:1241-56.
59. Wang J, Hu B, Cao F, Sun S, Zhang Y, Zhu Q. Down regulation of lncSCIR1 after spinal cord contusion injury in rat. Brain Res 2015;1624:314-20.
60. Zhou H, Shi Z, Kang Y, et al. Investigation of candidate long noncoding RNAs and messenger RNAs in the immediate phase of spinal cord injury based on gene expression profiles. Gene 2018;661:119-25.
61. Wang W, Su Y, Tang S, et al. Identification of noncoding RNA expression profiles and regulatory interaction networks following traumatic spinal cord injury by sequence analysis. Aging 2019;11:2352-68.
62. Moonga J, Likupe G. A systematic literature review on nurses’ and health care support workers’ experiences of caring for people with dementia on orthopaedic wards. J Clin Nurs 2016;25:1789-804.
63. Näslund J, Haroutunian V, Mohs R, et al. Correlation between elevated levels of amyloid beta-peptide in the brain and cognitive decline. JAMA 2000;283:1571-7.
64. Vassar R, Kandalepas PC. The β-secretase enzyme BACE1 as a therapeutic target for Alzheimer's disease. Alzheimers Res Ther 2011;3:20.
65. Riva P, Ratti A, Venturin M. The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr Alzheimer Res 2016;13:1219-31.
66. Airavaara M, Pletnikova O, Doyle ME, Zhang YE, Troncoso JC, Liu QR. Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J Biol Chem 2011;286:45093-102.
67. Massone S, Vassallo I, Fiorino G, et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis 2011;41:308-17.
68. Parenti R, Paratore S, Torrisi A, Cavallaro S. A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during beta-amyloid-induced apoptosis. Eur J Neurosci 2007;26:2444-57.
69. Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc Natl Acad Sci USA 2007;104:10679-84.
70. Ciarlo E, Massone S, Penna I, et al. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer's disease brain samples. Dis Model Mech 2013;6:424-33.
71. Massone S, Ciarlo E, Vella S, et al. NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid β secretion. Biochim Biophys Acta 2012;1823:1170-7.
72. Gu C, Chen C, Wu R, et al. Long noncoding RNA EBF3-AS promotes neuron apoptosis in Alzheimer's disease. DNA Cell Biol 2018;37:220-6.
73. Ferreira JJ, Rodrigues FB, Duarte GS, et al. An MDS evidence-based review on treatments for Huntington's disease. Mov Disord 2022;37:25-35.
74. Lee M, Liu T, Im W, Kim M. Exosomes from adipose-derived stem cells ameliorate phenotype of Huntington's disease in vitro model. Eur J Neurosci 2016;44:2114-9.
76. Chung DW, Rudnicki DD, Yu L, Margolis RL. A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Hum Mol Genet 2011;20:3467-77.
77. Sunwoo JS, Lee ST, Im W, et al. Altered expression of the long noncoding RNA NEAT1 in Huntington's disease. Mol Neurobiol 2017;54:1577-86.
78. Koistinen H, Kovanen RM, Hollenberg MD, et al. The roles of proteases in prostate cancer. IUBMB Life 2023;75:493-513.
79. Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 2014;5:5383.
80. Yuan S, Bi J, Zhang Y. LncRNA SSTR5-AS1 as a prognostic marker promotes cell proliferation and epithelial-to-mesenchymal transition in prostate cancer. Crit Rev Eukaryot Gene Expr 2023;33:1-12.
81. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33.
82. Sideris N, Dama P, Bayraktar S, Stiff T, Castellano L. LncRNAs in breast cancer: a link to future approaches. Cancer Gene Ther 2022;29:1866-77.
83. Sang Y, Tang J, Li S, et al. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16(INK4A) expression. Sci Rep 2016;6:22366.
84. Zhang J, Sui S, Wu H, et al. The transcriptional landscape of lncRNAs reveals the oncogenic function of LINC00511 in ER-negative breast cancer. Cell Death Dis 2019;10:599.
85. Li H, Chen C, Fan J, et al. Identification of cardiac long non-coding RNA profile in human dilated cardiomyopathy. Cardiovasc Res 2018;114:747-58.
86. Zhang X, Yuan S, Liu J, et al. Overexpression of cytosolic long noncoding RNA cytb protects against pressure-overload-induced heart failure via sponging microRNA-103-3p. Mol Ther Nucleic Acids 2022;27:1127-45.
87. Xie R, Fan J, Wen J, et al. LncRNA ZNF593-AS alleviates diabetic cardiomyopathy via suppressing IRF3 signaling pathway. Mol Ther Nucleic Acids 2023;32:689-703.
88. Fan J, Li H, Xie R, et al. LncRNA ZNF593-AS alleviates contractile dysfunction in dilated cardiomyopathy. Circ Res 2021;128:1708-23.
89. Zhong J, Jiang L, Huang Z, et al. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav Immun 2017;65:183-94.
90. Patel NA, Moss LD, Lee JY, et al. Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J Neuroinflamm 2018;15:204.
91. Harris L, Zalucki O, Piper M, Heng JI. Insights into the biology and therapeutic applications of neural stem cells. Stem Cells Int 2016;2016:9745315.
92. Zheng J, Yi D, Liu Y, Wang M, Zhu Y, Shi H. Long nonding RNA UCA1 regulates neural stem cell differentiation by controlling miR-1/Hes1 expression. Am J Transl Res 2017;9:3696-704. Available from: https://europepmc.org/article/MED/28861160 [Last accessed on 1 Apr 2024].
93. Liu Y, Pan L, Jiang A, Yin M. Hydrogen sulfide upregulated lncRNA CasC7 to reduce neuronal cell apoptosis in spinal cord ischemia-reperfusion injury rat. Biomed Pharmacother 2018;98:856-62.
94. Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev 2003;67:657-85.
95. Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 2008;14:723-30.
96. Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol Dis 2012;46:245-54.
97. Ksiazek-Winiarek DJ, Szpakowski P, Glabinski A. Neural plasticity in multiple sclerosis: the functional and molecular background. Neural Plast 2015;2015:307175.
98. Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci 2004;24:4250-8.
99. Zuccato C, Tartari M, Crotti A, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet 2003;35:76-83.
100. Ghafouri-Fard S, Khoshbakht T, Taheri M, Ghanbari M. A concise review on the role of BDNF-AS in human disorders. Biomed Pharmacother 2021;142:112051.
101. Yin X, Wang S, Ge R, et al. Long non-coding RNA DNMBP-AS1 promotes prostate cancer development by regulating LCLAT1. Syst Biol Reprod Med 2023;69:142-52.
102. Zhang W, Shi C, Xu Q, Chen X, Zhu H, Zheng B. Long non-coding RNA MIR22HG suppresses cell proliferation and promotes apoptosis in prostate cancer cells by sponging microRNA-9-3p. Bioengineered 2022;13:13108-17.
103. Ren Y, Jia HH, Xu YQ, et al. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ß1 secretion. Mol Cancer 2018;17:5.
104. Khorkova O, Wahlestedt C. Oligonucleotide therapies for disorders of the nervous system. Nat Biotechnol 2017;35:249-63.
105. Zhu Y, Zhu L, Wang X, Jin H. RNA-based therapeutics: an overview and prospectus. Cell Death Dis 2022;13:644.
106. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov 2021;20:629-51.
107. Chi X, Gatti P, Papoian T. Safety of antisense oligonucleotide and siRNA-based therapeutics. Drug Discov Today 2017;22:823-33.
108. Migliorati JM, Liu S, Liu A, et al. Absorption, distribution, metabolism, and excretion of US food and drug administration-approved antisense oligonucleotide drugs. Drug Metab Dispos 2022;50:888-97.
109. Germain ND, Chung WK, Sarmiere PD. RNA interference (RNAi)-based therapeutics for treatment of rare neurologic diseases. Mol Aspects Med 2023;91:101148.
110. Sheng P, Flood KA, Xie M. Short hairpin RNAs for strand-specific small interfering RNA production. Front Bioeng Biotechnol 2020;8:940.
111. Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med 2014;6:851-64.
112. Lima JF, Cerqueira L, Figueiredo C, Oliveira C, Azevedo NF. Anti-miRNA oligonucleotides: a comprehensive guide for design. RNA Biol 2018;15:338-52.
113. Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, Halsema N, Kroesen BJ, van den Berg A. Generation of miRNA sponge constructs. Methods 2012;58:113-7.
114. Chang S. Construction of multi-potent microRNA sponge and its functional evaluation. In: Wu W, editor. MicroRNA and cancer. New York: Springer; 2018. pp. 201-9.
115. Jung J, Yeom C, Choi YS, et al. Simultaneous inhibition of multiple oncogenic miRNAs by a multi-potent microRNA sponge. Oncotarget 2015;6:20370-87.
116. Das S, Kohr M, Dunkerly-Eyring B, et al. Divergent effects of miR-181 family members on myocardial function through protective cytosolic and detrimental mitochondrial microRNA targets. J Am Heart Assoc 2017;6:e004694.
117. Wang Z. The Principles of MiRNA-masking antisense oligonucleotides technology. In: Wu W, editor. MicroRNA and cancer. Totowa: Humana Press; 2011. pp. 43-9.
118. Gilot D, Migault M, Bachelot L, et al. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat Cell Biol 2017;19:1348-57.
119. Almarghalani DA, Boddu SHS, Ali M, et al. Small interfering RNAs based therapies for intracerebral hemorrhage: challenges and progress in drug delivery systems. Neural Regen Res 2022;17:1717-25.
120. Friedrich M, Aigner A. Therapeutic siRNA: state-of-the-art and future perspectives. BioDrugs 2022;36:549-71.
121. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96-118.
122. Modarresi F, Faghihi MA, Lopez-Toledano MA, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 2012;30:453-9.
123. Lee D, Chen W, Kaku HN, et al. Antisense oligonucleotide therapy rescues disturbed brain rhythms and sleep in juvenile and adult mouse models of Angelman syndrome. Elife 2023;12:e81892.
124. de Smet MD, Meenken CJ, van den Horn GJ. Fomivirsen - a phosphorothioate oligonucleotide for the treatment of CMV retinitis. Ocul Immunol Inflamm 1999;7:189-98.
125. Parham JS, Goldberg AC. Mipomersen and its use in familial hypercholesterolemia. Expert Opin Pharmaco 2019;20:127-31.
127. Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res 2014;42:609-21.
128. Gallant-Behm CL, Piper J, Lynch JM, et al. A MicroRNA-29 mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J Invest Dermatol 2019;139:1073-81.
129. Regulus Therapeutics I. RG-125 (AZD4076), a microRNA therapeutic targeting microRNA-103/107 for the treatment of NASH in patients with type 2 diabetes/pre-diabetes, selected as clinical candidate by AstraZeneca. 2015. Available from: https://www.prnewswire.com/news-releases/rg-125-azd4076-a-microrna-therapeutic-targeting-microrna-103107-for-the-treatment-of-nash-in-patients-with-type-2-diabetespre-diabetes-selected-as-clinical-candidate-by-astrazeneca-300062261.html [Last accessed on 1 Apr 2024].
131. Siddiqui MA, Keating GM. Pegaptanib: in exudative age-related macular degeneration. Drugs 2005;65:1571-7; discussion 1578-9.
132. Goodkey K, Aslesh T, Maruyama R, Yokota T. Nusinersen in the treatment of spinal muscular atrophy. In: Yokota T, Maruyama R, editors. Exon skipping and inclusion therapies. New York: Springer; 2018. pp. 69-76.
135. Bravo-Hernandez M, Tadokoro T, Navarro MR, et al. Spinal subpial delivery of AAV9 enables widespread gene silencing and blocks motoneuron degeneration in ALS. Nat Med 2020;26:118-30.
136. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med 2018;24:257-77.
138. Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 2016;44:863-77.
139. Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep 2014;6:211-21.
140. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol 2011;30:16-34.
141. Sioud M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2'-hydroxyl uridines in immune responses. Eur J Immunol 2006;36:1222-30.
143. Hong DS, Kang YK, Borad M, et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer 2020;122:1630-7.
144. Beg MS, Brenner AJ, Sachdev J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 2017;35:180-8.
145. Pisano M, Baldinu P, Sini MC, Ascierto PA, Tanda F, Palmieri G. Targeting Bcl-2 protein in treatment of melanoma still requires further clarifications. Ann Oncol 2008;19:2092-3.
146. Anderson EM, Miller P, Ilsley D, et al. Gene profiling study of G3139- and Bcl-2-targeting siRNAs identifies a unique G3139 molecular signature. Cancer Gene Ther 2006;13:406-14.
147. Winkler J, Stessl M, Amartey J, Noe CR. Off-target effects related to the phosphorothioate modification of nucleic acids. ChemMedChem 2010;5:1344-52.
148. Wen J, Friedman JR. miR-122 regulates hepatic lipid metabolism and tumor suppression. J Clin Invest 2012;122:2773-6.
149. Ui-Tei K, Naito Y, Nishi K, Juni A, Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res 2008;36:7100-9.
150. Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006;441:537-41.
151. Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 2009;27:549-55.
152. Hart M, Nickl L, Walch-Rueckheim B, et al. Wrinkle in the plan: miR-34a-5p impacts chemokine signaling by modulating CXCL10/CXCL11/CXCR3-axis in CD4+, CD8+ T cells, and M1 macrophages. J Immunother Cancer 2020;8:e001617.
153. LaCasse EC. Pulling the plug on a cancer cell by eliminating XIAP with AEG35156. Cancer Lett 2013;332:215-24.
154. Fazaeli H, Sheikholeslami A, Ghasemian F, Amini E, Sheykhhasan M. The emerging role of LncRNA FENDRR in multiple cancers: a review. Curr Mol Med 2023;23:606-29.
155. da Silva AMG, Cruz MS, de Souza KSC, Silbiger VN. Long non-coding RNA and circular RNA: new perspectives for molecular pathophysiology of atrial fibrillation. Mol Biol Rep 2023;50:2835-45.
156. Loganathan T, Doss C GP. Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets. Funct Integr Genomics 2023;23:33.
157. Jiapaer Z, Li C, Yang X, et al. Extracellular non-coding RNAs in cardiovascular diseases. Pharmaceutics 2023;15:155.
158. Dong B, Zhang F, Zhang W, Gao Y. IncRNA EPB41L4A-AS1 mitigates the proliferation of non-small-cell lung cancer cells through the miR-105-5p/GIMAP6 axis. Crit Rev Eukaryot Gene Expr 2023;33:27-40.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Kamal A, Swellam M, Shalaby NM, Darwish MK, El-Nahrery EM. Silent players, loud impact: unveiling the therapeutic potentials of LncRNAs. J Transl Genet Genom 2024;8:162-85. http://dx.doi.org/10.20517/jtgg.2023.55
AMA Style
Kamal A, Swellam M, Shalaby NM, Darwish MK, El-Nahrery EM. Silent players, loud impact: unveiling the therapeutic potentials of LncRNAs. Journal of Translational Genetics and Genomics. 2024; 8(2): 162-85. http://dx.doi.org/10.20517/jtgg.2023.55
Chicago/Turabian Style
Kamal, Ahmed, Menha Swellam, Nevin M. Shalaby, Marwa K. Darwish, Eslam M. El-Nahrery. 2024. "Silent players, loud impact: unveiling the therapeutic potentials of LncRNAs" Journal of Translational Genetics and Genomics. 8, no.2: 162-85. http://dx.doi.org/10.20517/jtgg.2023.55
ACS Style
Kamal, A.; Swellam M.; Shalaby NM.; Darwish MK.; El-Nahrery EM. Silent players, loud impact: unveiling the therapeutic potentials of LncRNAs. J. Transl. Genet. Genom. 2024, 8, 162-85. http://dx.doi.org/10.20517/jtgg.2023.55
About This Article
Special Issue
Copyright
Data & Comments
Data

 Cite This Article 0 clicks
Cite This Article 0 clicks




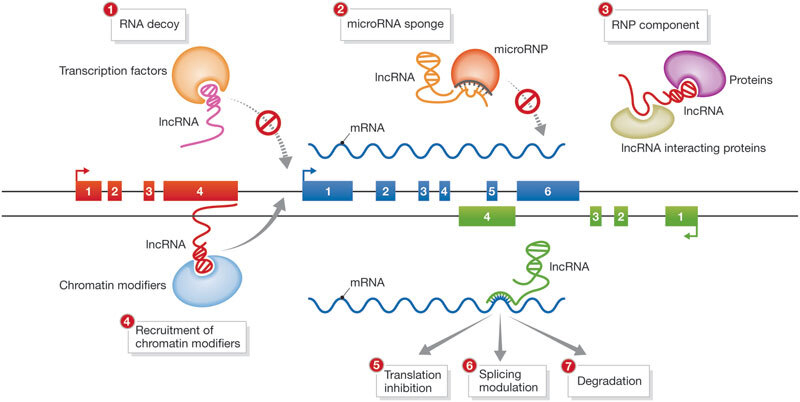
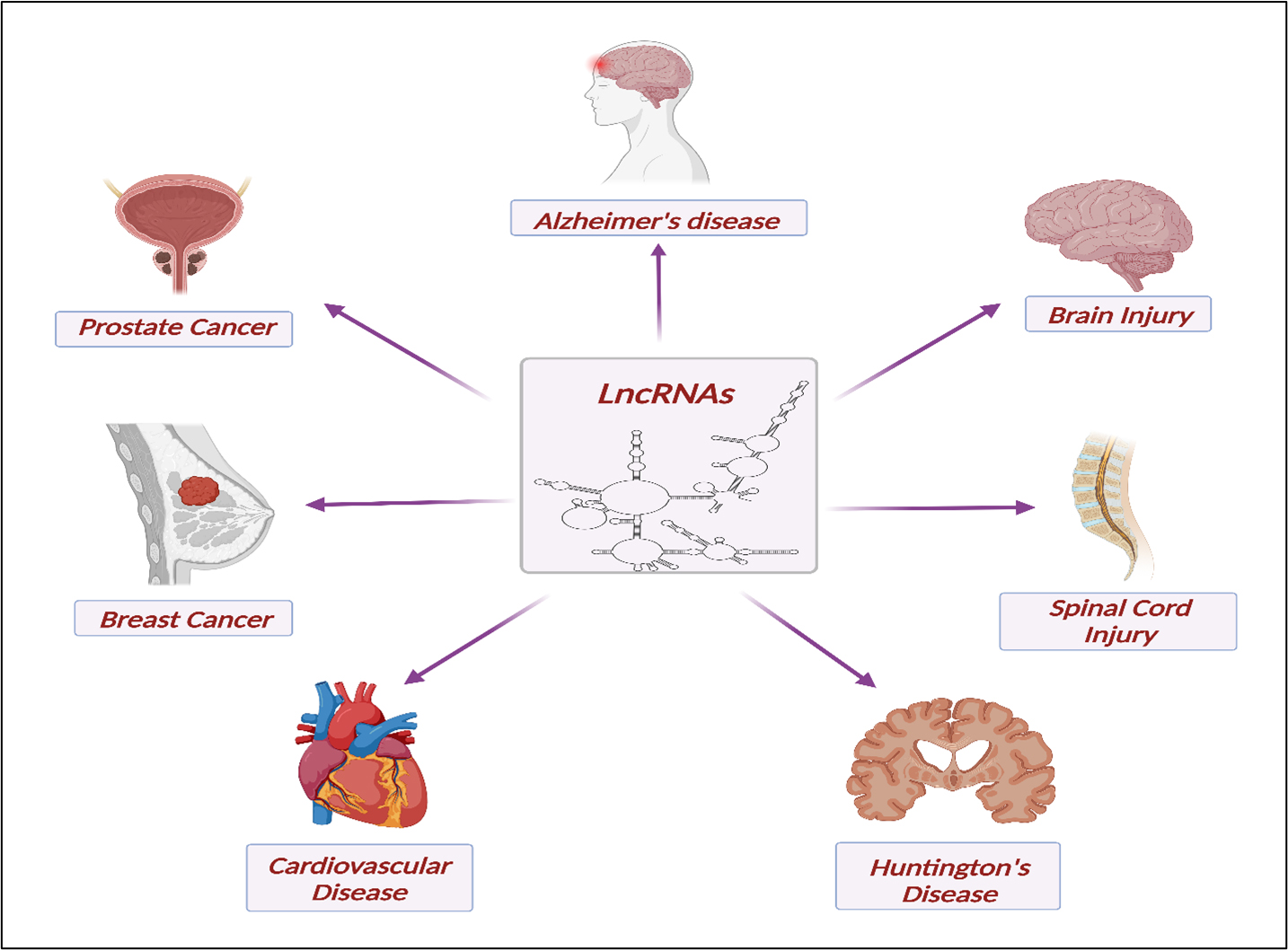
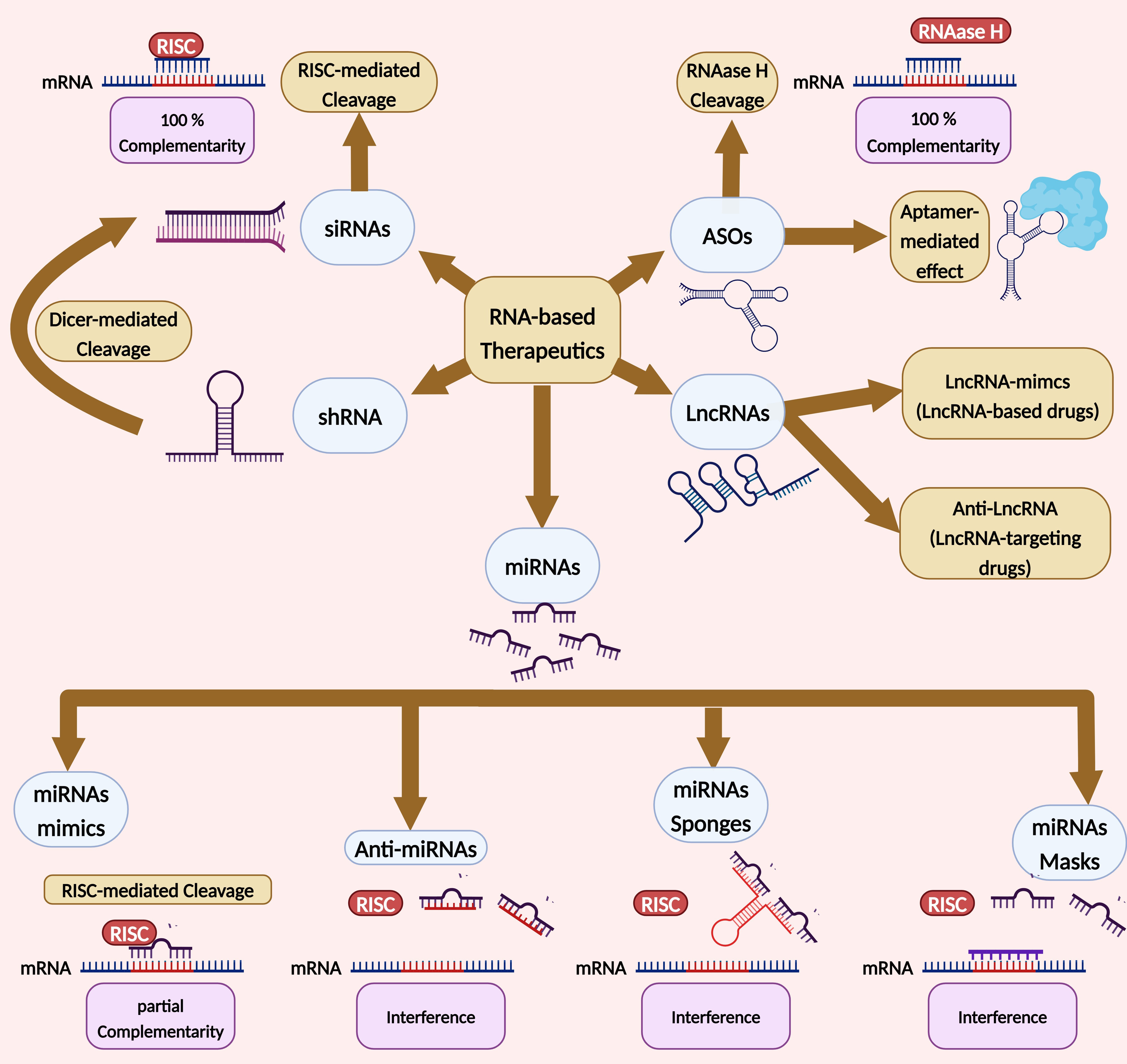








Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.