Individual xenografts for personalized treatment of women with metastatic triple negative breast cancer
Abstract
Aim: Triple negative breast cancer (TNBC) is the most severe subtype of breast cancer with poor prognosis even when treated at a localized stage. The treatment of metastatic TNBC is still challenging daily clinical practice, mainly because of the lack of targeted therapies. In the last years, a molecular sub-classification of TNBC has opened the way to personalized medicine for this type of severe cancer.
Methods: In this study, we assessed the added value of combining molecular analyses with individual xenografts to personalize the treatment of resort for five women with metastatic TNBC. While a patient was receiving one or two lines of chemotherapy, the corresponding xenograft model was tested with different drugs or drug combinations, mainly based on transcriptomic analyses of the tumor and on theoretical activated canonical pathways.
Results: On the basis of transcriptomic analyses and chemosensitivity data obtained from TNBC individual xenografts, we personalized the resort treatment for the five women in our study. In all cases, despite the fact that this resort treatment was a third-line or a fourth-line treatment, the time to progression was longer than that observed with previous lines of chemotherapy. When we explored the 19 chemotherapy regimens given to these women and their corresponding xenograft models, there was a strong correlation between ΔSUVmax (maximum standard uptake value) on positron emission tomography-computed tomography and the corresponding coefficients of inhibition obtained in mice.
Conclusion: The combination of gene expression profiling and individual xenografts is a promising method and could be proposed as a personalized therapeutic resort for women with metastatic TNBCs.
Keywords
Introduction
Triple negative breast cancer (TNBC) is the most severe subtype of breast cancer, occurring in younger women and associated with poor prognosis even when treated at a localized stage. TNBCs lack any detectable expression of estrogen receptor, progesterone receptor, or human epithelial growth factor receptor 2 (HER2) amplification. Treatment of metastatic TNBC is still challenging daily clinical practice, mainly because of a lack of targeted therapies.
At the end of the 19th century, Beatson[1] opened the way to hormone therapy and personalized medicine in oncology with the first successful “surgical castration” in a young pre-menopausal woman with metastatic breast cancer. In the late 1990s, an anti-HER2 monoclonal antibody, trastuzumab, was approved for the treatment of HER2 overexpressing metastatic breast cancer[2]. To date, 48 targeted therapies have been approved for the treatment of various types of metastatic cancers[3].
High-speed whole-genome sequencing technologies have led to the identification of numerous potential molecular targets. In 2000, a first molecular classification of breast cancer individualized four subtypes: luminal A, luminal B, HER2-enriched, and basal-like (BL)[4-6], most BL subtypes corresponding to triple-negative breast cancers[7,8]. In 2011, TNBCs were classified into 6 molecular subtypes including BL1, BL2, immunomodulatory, mesenchymal-like, mesenchymal stem-like, and luminal androgen receptor (LAR)[9]. According to this classification, BL1 tumors are likely to be more sensitive to cisplatin and DNA repair inhibitors, BL2 tumors to anti-epidermal growth factor receptor (EGFR) therapies, and LAR tumors to androgen inhibitors[10-12].
Patient-derived xenografts (PDXs) are rediscovered pre-clinical pharmacological models. They enable limited available quantities of human cancer tissue to be amplified. In addition, PDX models efficiently reflect the characteristics of the original cancer, including tumor heterogeneity and metastatic potential[13-15]. As a result, there is an excellent correlation between the anti-tumor activity of a given drug in a xenograft model, and the anti-tumor effect of the same drug in the corresponding patient[16-19].
In this pilot study, we combined individual PDXs and whole-genome analyses to personalize the resort treatment for women with metastatic TNBCs.
Methods
Patients with metastatic TNBC and gene expression profiling
Five women with metastatic TNBC participated in this study. For each patient, five tumor samples were obtained during an imagery-guided biopsy, at the time of metastatic disease, before any medical treatment. Informed written consent was obtained from the patients. The Clinical Research Board Ethics Committee (Comité de Protection des Personnes) approved this study (CPP Ile-de-France N°13218). Among these five tumor samples, (1) two were formaldehyde-fixed and paraffin-embedded for histological analyses; (2) two were immediately snap-frozen in liquid nitrogen and stored in the Hôpital-Saint-Louis tumor bank for molecular analysis; and (3) one was set aside in culture medium for xenografting.
Total RNA was extracted from the frozen tumor sample, and transcriptomic analyses were performed using a MiltenyiBiotec Microarray. A linear T7-based amplification step was performed from 0.5 µg of all RNA samples. To produce Cy3-labeled cRNA, the RNA samples were amplified and labeled using the Agilent-quick-labeling kit. The yields of cRNA and the dye-incorporation rate were measured on an ND-1000 spectrophotometer (NanoDrop, LabTech, France). Hybridization was performed according to the Agilent 60-mer oligo-microarray processing protocol: 1.65 µg Cy3-labeled cRNA was hybridized overnight at 65 °C on Agilent-Whole-Human-Genome-Oligo-Microarrays 4x44K, and fluorescence signals were detected using Agilent’s Microarray-Scanner. Agilent-FE-Software determined feature intensities and quantile normalization was performed with the Agi4x44PreProcess R package. Subsequent analyses were carried out with R3.01 software (Foundation for Statistical Computing, Vienna, Austria) and based on log2 single-intensity expression data. The classification was obtained by correlating gene expression profiles with the centroids for each of the 6 TNBC subtypes described by Lehmann et al.[9], and with Parker et al.[20] centroids for the PAM50 classification.
Patient-derived breast cancer xenografts and treatments
After imaging-guided tumor biopsies had been performed, one sample was transported in RPMI-1640 culture medium and subcutaneously grafted in 6-week-old NMRI-nude mice, under xylasin (10 mg/kg)/ketamin (100 mg/kg) anesthesia.
Four PDX models were successfully obtained, and studied [patient-derived xenograft of breast cancer 1 (PDXB1) to PDXB3 and PDXB5] [Table 1].
Patients’ characteristics at the time of transcriptomic analyses and of xenografting
| Patients | Age (year) | TNM | Metastases | Site of biopsy | Classification | Canonical pathways | Engraf-tments | |
|---|---|---|---|---|---|---|---|---|
| PAM50[20] | Lehmann 2011[9] | |||||||
| Patient 1 | 45 | T2N2M1 | Lung, brain, lymph nodes | Breast | Basal | BL2 | EGF pathway | PDXB1 |
| Patient 2 | 66 | T1N0M1 | Lung, liver, bone, lymph nodes | Lymph node | Basal | Stm | IGF/mTOR pathway | PDXB2 |
| Patient 3 | 30 | T4cN2M1 | Lung, lymph node | Breast | Basal | BL1 | Cell cycle/ BRCA | PDXB3 |
| Patient 4 | 40 | T2N1M1 | Lung, liver | Liver | Luminal B | AR | AR pathway | No |
| Patient 5 | 65 | T4dN1M1 | Lung, liver, bone, lymph nodes | Breast | Basal | BL2 | EGF pathway | PDXB5 |
For each xenograft model, after successful engraftment, a clinical score was recorded daily for the mice and tumor growth was measured in two perpendicular diameters with a caliper. Tumor volumes were calculated as V = L × l2 ÷ 2, L being the larger diameter (length), l the smaller (width). After mouse euthanasia, the tumor was resected, cut into small pieces of 1 mm3, and grafted again in 30 other nude mice. When tumors reached a volume of 300 mm3 (n = 4 mice per treatment-group), after 2 to 4 weeks, the mice were treated for one month with different regimens of chemotherapy [Table 2]. A daily clinical score was recorded and tumor growth measured weekly until its weight reached the ethically recommended limit of less than 10% of mouse weight (Directive 2010/63/EU of the European Parliament and the Council of 22 September 2010 on the protection of animals used for scientific purposes; Official Journal of the European Union L 276/33).
Administration of drugs in xenografted mice
| Drug | Dose | Mode of administration | Frequency of administration |
|---|---|---|---|
| Epirubicin | 1 mg/kg | Intra-peritoneal | Once a week |
| Cyclophosphamide | 30 mg/kg | Intra-peritoneal | Twice a week |
| Bevacizumab | 5 mg/kg | Intra-peritoneal | Three times a week |
| Paclitaxel | 20 mg/kg | Intra-peritoneal | Twice a week |
| Docetaxel | 15 mg/kg | Intra-peritoneal | Twice a week |
| Cetuximab | 10 mg/kg | Intra-peritoneal | Twice a week |
| Cisplatin | 3 mg/kg | Intra-peritoneal | Once a week |
| Oxaliplatin | 1 mg/kg | Intra-peritoneal | Twice a week |
| Gemcitabine | 400 mg/kg | Intra-peritoneal | Once a week |
| Capecitabine | 400 mg/kg | Gavage | Once a week |
| Everolimus | 5 mg/kg | Gavage | Twice a week |
| Sunitinib | 40 mg/kg | Gavage | Once a week |
Assessment of tumor response in patients
For each line of chemotherapy, the patient response under treatment was characterized. Metabolic response was assessed according to positron emission tomography (PET) response criteria in solid tumors criteria[21]. Briefly, partial metabolic response (PMR) is defined as a reduction in maximum standard uptake value (SUVmax) of at least 30%, with no new lesions. Complete metabolic response (CMR) corresponds to the disappearance of all lesions in the blood-pool background. Progressive metabolic disease (PMD) is defined by an increase in SUVmax greater than 30%, or the appearance of new fluoro-deoxyglucose (FDG)-avid lesions. Stable metabolic disease applies when the criteria for the other categories (CMR, PMR or PMD) are not met.
18FDG (5 MBq/kg; not exceeding 500 MBq) was injected intravenously 60 min before data were acquired on a Philips gemini XL PET/computed tomography (CT) scanner. CT data were acquired first (120 kV; 100 mAs; no contrast-enhancement). PET 3D data were acquired with 2 min per bed position, and images were reconstructed using a 3D row-action maximum likelihood algorithm.
PET/CT images were interpreted by a nuclear medicine physician blinded to the patient’s record. 18FDG uptake was expressed as the SUV. A 3D region of interest (ROI) was drawn around the lesions and SUVmax (maximum SUV value within the ROI) was measured. SUVmax of the lesions with the highest uptake were recorded and used for the study analysis (five target lesions were assessed). The SUVmax of the liver was also recorded as a control value. The change in SUVmax at each evaluation was expressed as ΔSUVmax (%) = 100 × (cycle n SUVmax - cycle (n - 1) SUVmax)/cycle (n - 1) SUVmax. The appearance of new lesions was also recorded.
For each line of chemotherapy, time-to-progression (TTP) was defined as the time between the initiation of treatment and the diagnosis of disease progression.
Assessment of tumor response in xenografts
To assess tumor response in xenografts, ultrasonography was performed twice a week on treated and untreated mice using an AplioXT ultrasonograph (Toshiba, Japan). Tumor growth was measured in two diameters, and tumor volumes were calculated as V = L × l2 ÷ 2. For each drug or drug combination, a growth curve was established.
The coefficient of inhibition for a drug or a drug combination was calculated as (a’- a)/a, a being the slope of the curve before the start of treatment (day 0), and a’ the slope of the curve between day 0 and day 28 of treatment [Figure 1].
Statistical analysis
For the correlation studies, the Kendall rank correlation coefficient R2 was calculated between patient ΔSUVmax for a given chemotherapy regimen and the coefficient of inhibition for the same regimen in the TNBC xenograft. A P value under 0.05 was considered to be significant.
Results
Molecular signature of metastatic TNBCs and corresponding xenografts
Five women with metastatic TNBC were included in this pilot study. Their characteristics are detailed in Table 1. With their consent, a tumor biopsy was performed at the time of metastatic disease, before any medical treatment. One biopsy sample was dedicated to transcriptomic analyses, enabling the tumors to be classified according to Lehmann’s classification. Another biopsy sample was immediately processed for xenografting in nude mice. The graft was successful in 4 of the 5 patients [Table 1]. The molecular signature of each xenograft model was identical to the corresponding TNBC it derived from, and remained unchanged over successive passages for each xenograft model.
Personalizing treatment of metastatic breast cancers
While a patient was receiving one or two lines of chemotherapy, the corresponding xenograft model was tested with different drugs or drug combinations, the choice being mainly based on theoretical activated pathways identified from transcriptomic analyses [Table 1].
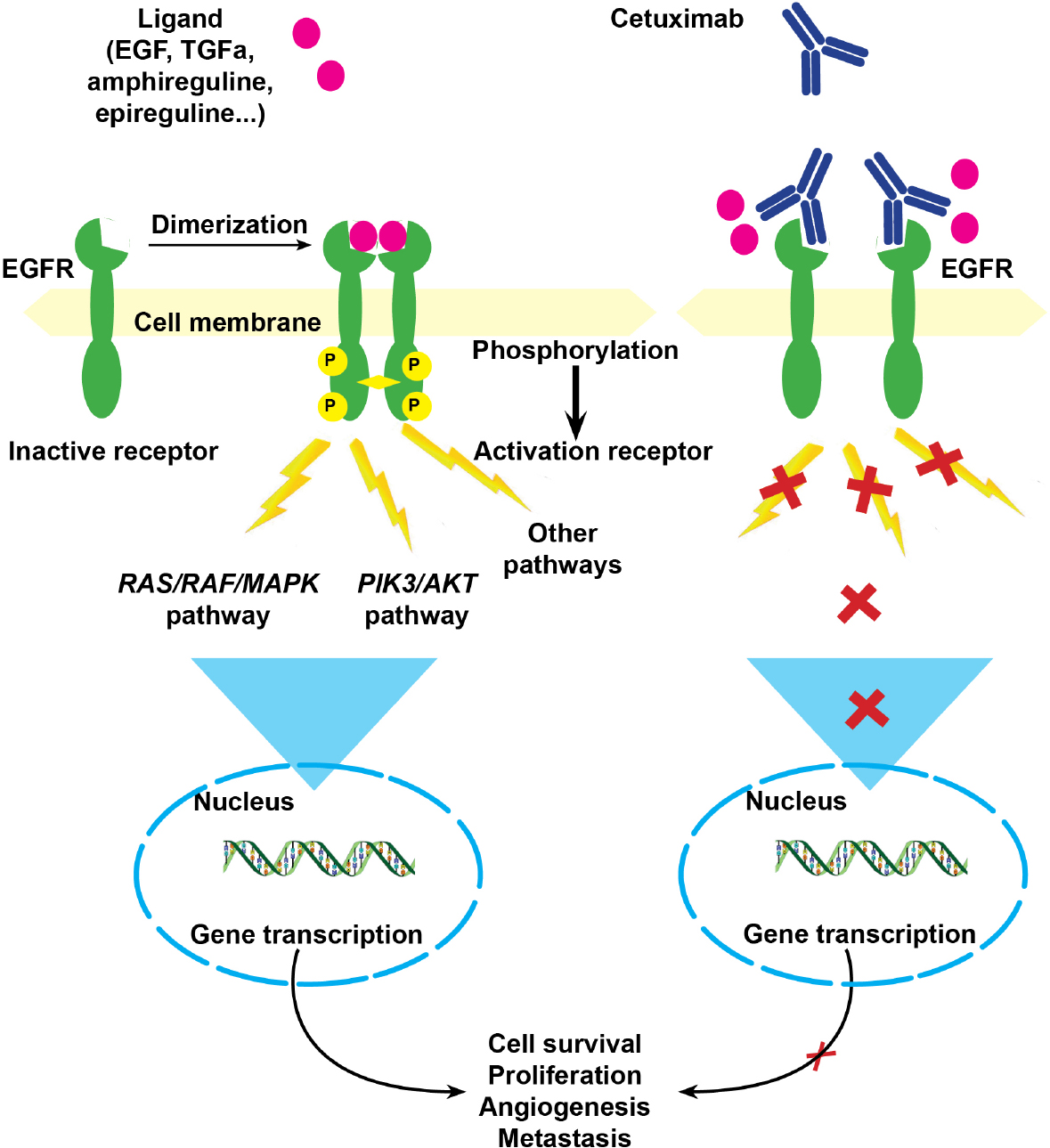
PDXB1, derived from the TNBC of patient 1, was classified BL2, with epidermal growth factor (EGF) pathway activation. In addition, there was no mutation of EGF pathway genes, mainly BRAF, KRAS, NRAS and PIK3. When tested with drugs or drug combinations on PDXB1, the most efficient regimen was a combination of paclitaxel and cetuximab, an anti-EGFR monoclonal antibody [Figure 2]. This regimen was then offered to the patient as a third-line resort treatment [Table 3]. Under this chemotherapy, she had almost CMR[13].
Figure 2. Epidermal growth factor pathway in triple negative breast cancers. EGFR is a tyrosine-kinase transmembrane receptor. When a ligand, mainly EGF, binds the extracellular domain, there is a dimerization of two receptors. This leads to their activation through phosphorylation of the intracellular tyrosine kinase. Cetuximab is a humanized anti-EGFR receptor, which competitively binds to the extracellular domain, blocking receptor dimerization. EGF: epidermal growth factor; EGFR: epidermal growth factor receptor; TGFα: transforming growth factor-α; MAPK: mitogen-activated protein kinase; PIK3: phosphatidylinositol 3-kinase
Response to treatment in patients and corresponding patient-derived xenografts
| Patients | Treatment | Best response in patients/ΔSUVmax | Coefficient of inhibition in corresponding PDX | TTP (months) |
|---|---|---|---|---|
| Patient 1 | Cisplatin gemcitabine | PMD/2.1 | 1.52 | 2 |
| Capecitabine bevacizumab | SMD/0.02 | -0.88 | 6 | |
| Paclitaxel cetuximab | PMR/-0.88 | -1.67 | 10 | |
| Patient 2 | Paclitaxel bevacizumab | PMD/0.31 | 1.57 | 3 |
| Capecitabin bevacizumab | PMD/0.41 | 1.2 | 3 | |
| Sunitinib | PMD/2.13 | 3.59 | 2 | |
| Cisplatin everolimus | PMR/-0.78 | -1.04 | 6 | |
| Patient 3 | SIM | PMR/-0.56 | -1.61 | 6 |
| Docetaxel | PMD/0.42 | 1.18 | 2 | |
| Cisplatin gemcitabine | SMD/0.03 | -1.42 | 8 | |
| Patient 4 | Palitaxel bevacizumab | PMR | NA | 12 |
| Cisplatin gemcitabine | PMD | NA | 2 | |
| Bicalutamide | SMD | NA | 8 | |
| Abiraterone acetate | PMR | NA | 10 | |
| Patient 5 | SIM | SMD/0.07 | 0.61 | 4 |
| Docetaxel | PMD/2.18 | 1.67 | 4 |
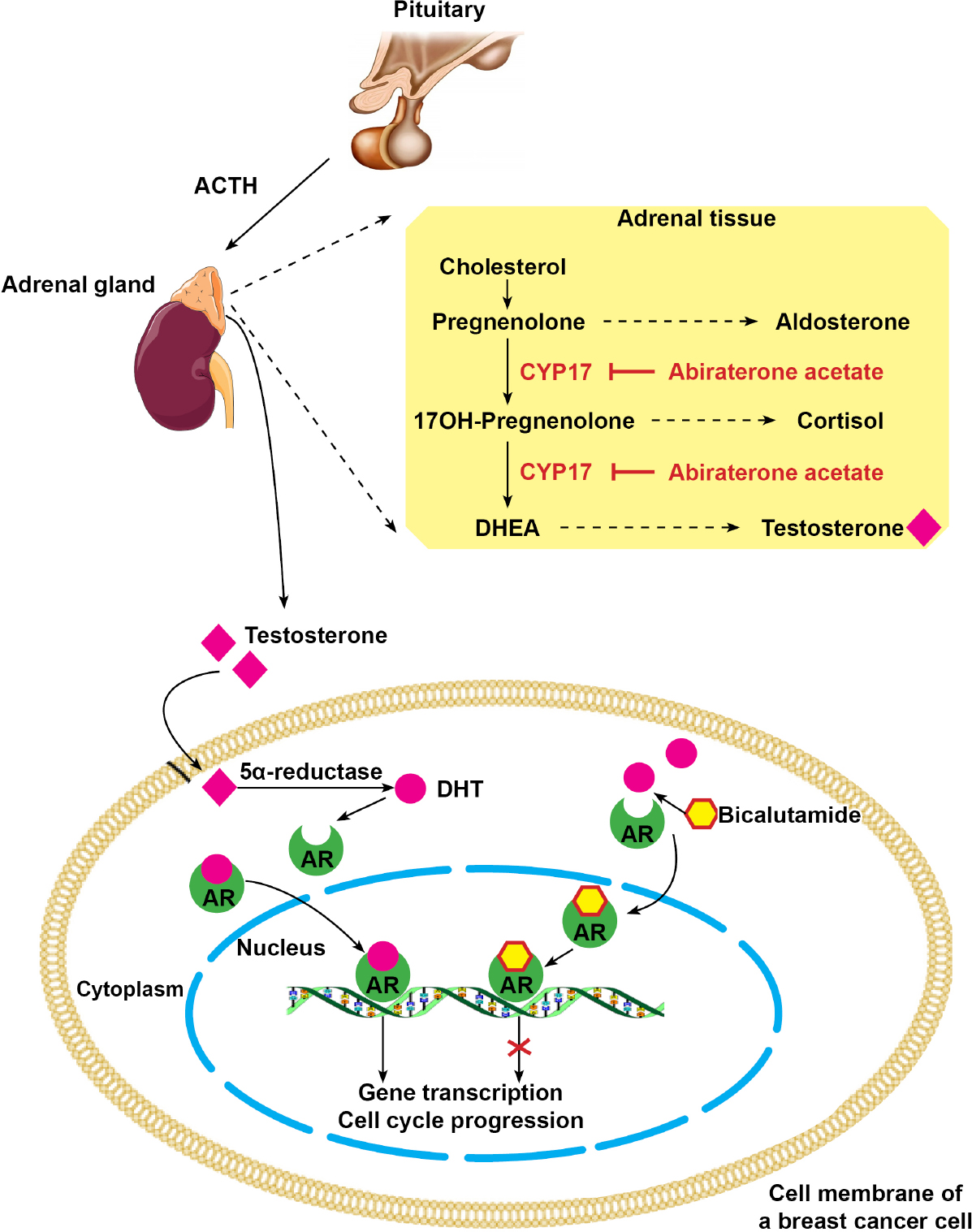
For patient 4, the engraftment was not successful. However, since the biopsied liver metastasis was classified as LAR, we decided to personalize the anti-cancer treatment using anti-androgen drugs. The patient was first offered a treatment with bicalutamide, an anti-androgen receptor, resulting in stable disease and an 8 months period to progression. She was then offered a second-line treatment with abiraterone acetate, a CYP17A inhibitor that blocks androgen production [Figure 3], which enabled an additional 10 months of disease control [Table 3].
Figure 3. Androgen pathway in triple negative breast cancers. Bicalutamide acts as an inhibitor competing with DHT on cytoplasmic AR. Abiraterone acetate inhibits CYP17, thus reducing testosterone synthesis in adrenal tissue. DHT: dihydrotestosterone; AR: androgen receptors; CYP17: cytochrome P450 17 alpha-hydroxylase; ACTH: adrenocorticotropic hormone; DHEA: dehydroepiandrosterone
Treatment efficacy in patients and in corresponding PDXs
For each of the four patients with a corresponding xenograft, we retrospectively analyzed response data for the different chemotherapy regimens they had received. In a given patient, for each regimen, we determined two parameters: the best change in SUVmax expressed as a percentage of SUVmax, and the time to progression. Each chemotherapy regimen was also tested in the corresponding TNBC xenograft, and tumor response was determined by the coefficient of inhibition. When we explored the 19 regimens given to the four patients and their corresponding xenograft models [Table 3], there was a strong correlation between ΔSUVmax and the corresponding coefficients of inhibition [Figure 4].
Figure 4. Correlation between inhibition coefficients and ΔSUVmax. For a drug or a drug combination, the inhibition coefficient is calculated as (a’-a)/a. The Kendall rank correlation coefficient is calculated between a given patient ΔSUVmax for one chemotherapy regimen and the inhibition coefficient for the same regimen in his corresponding TNBC xenograft. There is a significant correlation between these two parameters (R2 = 0.7, P < 0.05). TNBC: triple negative breast cancer; SUV: standard uptake value
On the basis of transcriptomic analyses and chemosensitivity data obtained from the different TNBC xenografts, we personalized resort treatment for the five women in our study. In all cases, despite the fact that this resort treatment was a third-line or a fourth-line, the TTP was longer than that observed with previous lines of chemotherapy [Table 3].
Discussion
TNBC is a heterogeneous, severe type of breast cancer, requiring the development of personalized therapies. Recent advances in gene expression profiling have identified TNBC molecular sub-types that could benefit from the use of targeted therapies[9].
Typically, BL2 subtype is characterized by an activation of the EGF pathway and could benefit from anti-EGFR therapies. In a phase II study on 173 women with metastatic TNBCs, where cetuximab was associated with cisplatin, it only added 2.2 months of survival[22]. However, the patients were not selected according to EGF pathway activation, which certainly lowered the benefit observed. In our pilot study, patient 1 had an activation of the EGF pathway with no mutation in the RAS, RAF or PIK3 genes, as observed in 40% of patients with metastatic colorectal cancer[23,24], where 60% response to anti-EGFR combined chemotherapy was observed[25]. Indeed, when patient 1 received a combination of paclitaxel and cetuximab as third-line treatment, she had almost a CMR.
On the other hand, LAR tumors account for 11% of TNBCs and are characterized by an activation of the androgen receptor pathway. In two phase-II studies on women with metastatic TNBCs selected according to androgen receptor status, bicalutamide or abiraterone acetate led to 6 months disease stabilization in less than 20% of patients[26,27]. Again, these disappointing results were probably due to an inadequate selection of patients. Indeed, TNBC tumors classified as LAR usually correspond to apocrine molecular tumors[28], and are more accurately identified using gross cystic disease fluid protein-15 marker by immunohistochemistry[29]. In our pilot study, the tumor of patient 4 was classified as LAR subtype, and this young woman drew durable benefit from anti-androgen therapies, with a total of 18 months of liver metastasis stabilization.
The molecular sub-classification of TNBCs by Lehman et al.[9] has opened the way to personalized medicine for metastatic TNBCs. However, molecular analyses still have limitations, and different, complementary methods need to be implemented. Individual xenografts from metastatic samples of TNBCs are an additional, innovative tool and are more physiological than genomic analyses. A major limitation of PDXs is the low engraftment rate. However, for metastatic TNBCs, the engraftment probability of metastatic samples is high, up to 80%[30]. Individual PDXs could thus be useful in co-clinical trials, using individual xenografts for the development of new therapeutic agents in patients with cancer resistant to standard treatments[13,31,32].
The combination of gene expression profiling and individual PDXs is a promising method, and could be proposed as a personalized therapeutic resort for women with metastatic TNBCs.
Declarations
AcknowledgmentsWe would like to thank Mrs. A. Swaine for revising the English language.
Authors’ contributionsCollected and assembled the data: Nguyen TT, Bousquet G
Provided data analysis and interpretation: Nguyen TT, El Bouchtaoui M, Janin A, Bousquet G
Drafted the manuscript: Nguyen TT, Janin A, Bousquet G
Analyzed the transcriptomic data: Feugeas JP
Provided the analysis of PET/CT data: Vercillino L
Performed imaging-guided tumor biopsies: de Bazelaire C
Located financial support and provided administrative support: Janin A
Conceived and designed the study, provided the study material or patients: Bousquet G
Read and approved the final manuscript: Nguyen TT, El Bouchtaoui M, Feugeas JP, Vercillino L, de Bazelaire C, Janin A, Bousquet G
Availability of data and materialsAll data generated or analyzed during this study are included in this published article and its supplementary information files.
Financial support and sponsorshipThe study was supported by INSERM and University Paris 7.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateThe Clinical Research Board Ethics Committee (Comité de Protection des Personnes) approved this study (CPP Ile-de-France N°13218). Informed written consent was obtained from the patients.
Consent for publicationWritten informed consent was obtained from the patients for publication.
Copyright© The Author(s) 2018.
REFERENCES
1. Beatson GT. On the treatment of inoperable cases of carcinoma of the mamma: suggestions for a new method of treatment, with illustrative cases. The Lancet 1896;148:162-5.
2. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783-92.
3. National Comprehensive Cancer Network. About the NCCN clinical practice guidelines in oncology (NCCN guidelines®). Available from: https://www.nccn.org/professionals. [Last accessed on 5 Nov 2018].
4. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52.
5. Trop I, LeBlanc SM, David J, Lalonde L, Tran-Thanh D, et al. Molecular classification of infiltrating breast cancer: toward personalized therapy. Radiographics 2014;34:1178-95.
6. Morigi C. Highlights from the 15th St Gallen International Breast Cancer Conference 15-18 March,. 2017, Vienna: tailored treatments for patients with early breast cancer. Ecancermedicalscience 2017;11:732.
7. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010;363:1938-48.
8. Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res 2007;9:R65.
9. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750-67.
10. Mancini P, Angeloni A, Risi E, Orsi E, Mezi S. Standard of care and promising new agents for triple negative metastatic breast cancer. Cancers (Basel) 2014;6:2187-223.
11. Nakai K, Hung MC, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res 2016;6:1609-23.
12. Tomao F, Papa A, Zaccarelli E, Rossi L, Caruso D, et al. Triple-negative breast cancer: new perspectives for targeted therapies. OncoTargets Ther 2015;8:177-93.
13. Bousquet G, Feugeas JP, Ferreira I, Vercellino L, Jourdan N, et al. Individual xenograft as a personalized therapeutic resort for women with metastatic triple-negative breast carcinoma. Breast Cancer Res 2014;16:401.
14. Reyal F, Guyader C, Decraene C, Lucchesi C, Auger N, et al. Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res 2012;14:R11.
15. Pompili L, Porru M, Caruso C, Biroccio A, Leonetti C. Patient-derived xenografts: a relevant preclinical model for drug development. J Exp Clin Cancer Res 2016;35:189.
16. Villarroel MC, Rajeshkumar NV, Garrido-Laguna I, De Jesus-Acosta A, Jones S, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther 2011;10:3-8.
17. Hidalgo M, Bruckheimer E, Rajeshkumar NV, Garrido-Laguna I, De Oliveira E, et al. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol Cancer Ther 2011;10:1311-6.
18. Tabernero J, Kunzmann V, Scheithauer W, Reni M, Shiansong Li J, et al. nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: a subgroup analysis of the Western European cohort of the MPACT trial. OncoTargets Ther 2017;10:591-6.
19. Bousquet G, Janin A. Patient-derived xenograft: an adjuvant technology for the treatment of metastatic disease. Pathobiology 2016;83:170-6.
20. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160-7.
21. O JH, Lodge MA, Wahl RL. Practical PERCIST: a simplified guide to PET response criteria in solid tumors 1.0. Radiology 2016;280:576-84.
22. Baselga J, Gómez P, Greil R, Braga S, Climent MA, et al. Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J Clin Oncol 2013;31:2586-92.
23. De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-62.
24. Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34.
25. Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27:2091-6.
26. Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic breast cancer. Clin Cancer Res 2013;19:5505-12.
27. Bonnefoi H, Grellety T, Tredan O, Saghatchian M, Dalenc F, et al. A phase II trial of abiraterone acetate plus prednisone in patients with triple-negative androgen receptor positive locally advanced or metastatic breast cancer (UCBG 12-1). Ann Oncol 2016;27:812-8.
28. Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 2005;24:4660-71.
29. Lehmann-Che J, Hamy AS, Porcher R, Barritault M, Bouhidel F, et al. Molecular apocrine breast cancers are aggressive estrogen receptor negative tumors overexpressing either HER2 or GCDFP15. Breast Cancer Res 2013;15:R37.
30. Gonçalves A, Bertucci F, Guille A, Garnier S, Adelaide J, et al. Targeted NGS, array-CGH, and patient-derived tumor xenografts for precision medicine in advanced breast cancer: a single-center prospective study. Oncotarget 2016;7:79428-41.
31. Misale S, Bozic I, Tong J, Peraza-Penton A, Lallo A, et al. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nat Commun 2015;6:8305.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Nguyen TT, El Bouchtaoui M, Feugeas JP, Vercillino L, de Bazelaire C, Janin A, Bousquet G. Individual xenografts for personalized treatment of women with metastatic triple negative breast cancer. J Transl Genet Genom 2018;2:19. http://dx.doi.org/10.20517/jtgg.2018.20
AMA Style
Nguyen TT, El Bouchtaoui M, Feugeas JP, Vercillino L, de Bazelaire C, Janin A, Bousquet G. Individual xenografts for personalized treatment of women with metastatic triple negative breast cancer. Journal of Translational Genetics and Genomics. 2018; 2: 19. http://dx.doi.org/10.20517/jtgg.2018.20
Chicago/Turabian Style
Nguyen, Thuy T., Morad El Bouchtaoui, Jean-Paul Feugeas, Laetitia Vercillino, Cédric de Bazelaire, Anne Janin, Guilhem Bousquet. 2018. "Individual xenografts for personalized treatment of women with metastatic triple negative breast cancer" Journal of Translational Genetics and Genomics. 2: 19. http://dx.doi.org/10.20517/jtgg.2018.20
ACS Style
Nguyen, TT.; El Bouchtaoui M.; Feugeas J.P.; Vercillino L.; de Bazelaire C.; Janin A.; Bousquet G. Individual xenografts for personalized treatment of women with metastatic triple negative breast cancer. J. Transl. Genet. Genom. 2018, 2, 19. http://dx.doi.org/10.20517/jtgg.2018.20
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 4 clicks
Cite This Article 4 clicks













Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.