Roles of gut dysbiosis, anti-proliferative proteins, and post-transcriptional regulation in carcinogenesis
Abstract
The potentially powerful impact of microbiota has attracted much attention. For example, dysbiosis of the gut microbiota could be linked to various cancers. It is probable that DNA damage and DNA repair impairment due to inflammation from gut dysbiosis would be of importance in carcinogenesis and/or preventing carcinogenesis. In fact, the signature of the gut microbiome has been shown to be associated with responses and/or successful survival rate to certain immune-blockade therapy in several cancers. Conversely, living cells have to cope with the danger of reactive oxygen species (ROS) disturbing the integrity of biomolecules, which can eventually lead to carcinogenesis if otherwise untreated. Gut microbiota could modulate considerable levels of ROS and oxidative damage. Interestingly, an anti-proliferative family (APRO) characterized by several immediate early responsive gene products might be deeply involved in the mechanism of carcinogenesis. It has been described that APRO proteins also participate in a variety of cellular processes including cell division, DNA repair, and mRNA stability. The biological function of APRO proteins seems to be quite complicated; however, they might be a key modulator of microRNAs (miRNAs) for post-transcriptional regulation. The next generation of therapy would likely contain strategies for modifying the redox background as well as the regulation of ROS in cells and/or for better DNA repair machinery with the APRO proteins via the modulation of miRNA-derived post-transcriptional regulation in a sustainable manner. Given the important function of the gut microbiota in balancing the immune network, carcinogenesis could therefore be prevented by suitable gut microbiota via the roles of APRO proteins. Consequently, probiotics might play a key role in the modulation of gut immune system in keeping healthy and/or preventing cancers.
Keywords
INTRODUCTION
Microbiota is microorganisms including bacteria living in the digestive tracts. The gut is the main location of human microbiota. The gut microbiota is held in an intricate balance[1]. Alterations in the microbiota are known to be influenced by several dietary, genetic, and environmental factors[2], which have been shown to trigger redox signaling within the gut mucosa[3]. Control of redox signaling may direct the outcome of an inflammatory event[4]. Therefore, gut microbiota transducing the redox signaling represents a performer in the control of the response to cancer therapies[5]. Redox medicine is a new therapeutic concept directing reactive oxygen species (ROS) for health improvement[6]. However, excessive ROS production causes oxidative stress, which is involved in the pathogenesis of various cancers[7]. While the production of ROS is a cause of DNA damage, the DNA damage could also be induced by alterations in the DNA repair machinery[8]. Gut microbiota is intensely involved in the signaling of ROS production, DNA damage, and DNA repair machinery within host cells. Therefore, inflammation, gut microbiota, ROS, and cancer therapeutics are closely associated with each other, which can affect the occurrence, progression, and treatment of cancers[9]. Several machineries in a cell are also responsible for multidrug resistance. It has been shown that complex redox signaling networks can regulate the pathways related to the progress of resistance[10]. The development of multidrug resistance generally characterizes the major problem of cancer therapy, which may result in poor prognosis of cancer patients[11]. Fundamental genetic modifications in cancer cells via several DNA damages by ROS may induce the changes of various genes’ expressions, regulating specific pathways that could control DNA repair, apoptosis, autophagy, and drug targets[12,13].
In our gut, there are numerous bacteria that are valuable to us, but pathogenic microorganisms may also exist. The commensal bacteria can even regulate the effectiveness of immune checkpoint cancer therapy by modulating the immune response to cancers[14]. For example, the benefits of therapy could be observed in cancer patients treated with anti-programmed cell death ligand 1 (PDL1), anti- Programmed cell death protein 1 (PD1), and/or anti-cytotoxic T-lymphocyte-associated protein 4 (CTLA4) blockade therapies. Microbial imbalance in the gut, called dysbiosis, is now supposed as one of the entrances to cancers[14,15]. In addition, the gut microbiota could increase the efficacy and decrease the toxicity of current chemotherapy mediators[16]. Communication between gut microbiota and host cells may signify a novel research area into potential machineries regulating the efficacy of cancer immune therapies. Especially, host-directed immunomodulation via the gut microbiota appears encouraging for a successful defense against cancers. Here, we summarize the key roles of ROS and/or inflammation in homeostasis, gut microbiota, and interesting anti-proliferative (APRO) family family proteins involved in the cell signaling, which would address emerging therapeutic strategies with the potential molecular mechanisms to strengthen protective host immunity against cancers.
GUT-CANCER AXIS IN CARCINOGENESIS
Gut microbiota consists of a multispecies microbial community that develops symbiosis with the host organism[17]. The microbiota is an important source of various metabolites, which exert important functions on the regulation of intestinal epithelial and immune homeostasis[18]. Gut dysbiosis is a perturbation of the microbiota, which may disturb the structure and/or function of the gut proteome and gut immunity[19]. Therefore, dysbiosis could lead to many diseases such as inflammatory bowel disease and/or cancer [Figure 1]. In fact, several studies have revealed an association between poor gut microbiota and carcinogenesis. For example, close relationships between the gut microbiota and colorectal cancer in humans have been found, which are implicated in intricate immune responses of gut dysbiosis influencing carcinogenesis[20]. Among gut microbiota-derived metabolites, trimethylamine-N-oxide has a possible role in the carcinogenesis of colorectal cancer[21]. However, the specific stimulus of the metabolites on carcinogenesis may be conditional. As mentioned above, diet has an influence on the construction and/or metabolism of the gut microbiota. Several metabolites, such as short-chain fatty acids (SCFAs) and indole derivatives, may act as guardians against carcinogenesis by adjusting immune responses[20]. The gut microbiome has also been linked to gastric and esophageal cancers[22]. In addition, gut dysbiosis is associated with a cycle of hepatocyte injury and regeneration characteristic of chronic liver disease, suggesting that the gut microbiota influences hepatic carcinogenesis[23]. Actually, it has been shown that the gut microbiota is associated with the development of hepatocellular carcinoma[24]. Different features of the gut microbiota have been observed between prostate cancer patients and benign prostatic hypertrophy, while the former has presented a higher prevalence of Bacteroides massiliensis compared to cases of benign prostatic hypertrophy[25]. Furthermore, the fecal microbiota of breast cancer and prostate cancer patients differs from that of control individuals[26]. In postmenopausal breast cancer patients, an altered fecal microbiota composition has also been observed compared with healthy controls[27]. Of note, alterations of the gut microbiota have been associated with increased estrogen deconjugation through the secretion of β-glucuronidase, suggesting the possibility of estrogen-related malignancy developing due to microbiota alteration[28]. In addition, gut microbiota closely participates in the development of lung cancer[29]. Furthermore, a supportive role for the gut microbiota has been shown in the development of lymphoid neoplasms[30]. DNA of Helicobacter spp. has been frequently reported to be detected in pancreatic cancer tissues[31]. Therefore, Helicobacter pylori infection may be a considerable risk factor for pancreatic ductal adenocarcinoma[32]. In addition, Helicobacter spp. is thought to be involved in chronic and/or acute pancreatitis as well as autoimmune pancreatitis[33]. An increase in these bacteria species is also associated with bile duct inflammation, the severity of bile duct fibrosis, and cholangiocarcinoma proliferation[34]. Conversely, it has been shown that metformin significantly changes the composition and/or predicted function of the gut microbiota in mice, suggesting an implication of metformin usage in the prevention of certain cancers[35]. Thus, many studies have pointed toward the gut microbiota as a key manager in host conditions against the development of various cancers. In other words, microbial imbalance and/or dysbiosis are now supposed as one of the gateways to several cancers.
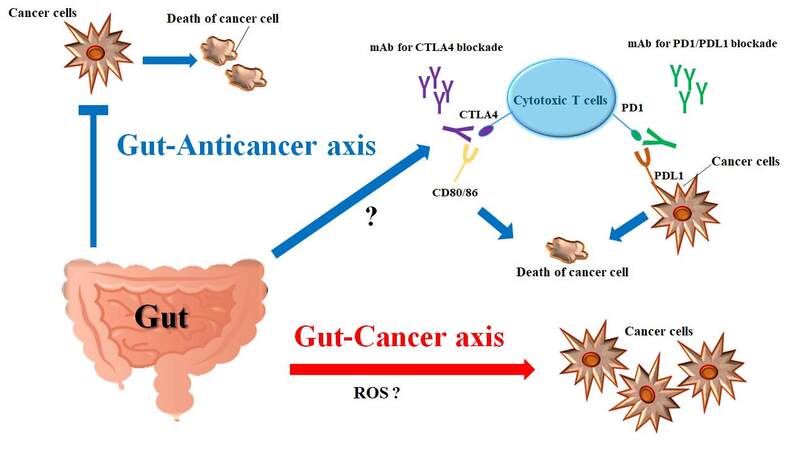
Figure 1. Illustration of the relationship among gut microbiota, cytotoxic T cells, cancer cells, and immune checkpoints inhibitors. There might be two opposite actions of gut microbiota. One is oncogenic or the cancer-causing gut-cancer axis. The other is the gut-anticancer axis favorable to the host. Immune checkpoint molecules are involved in the cell-cell communications, which are present on T cells and shared on cancer cells. Immune checkpoint molecules send signals to inhibit T cell activities of killing cancer cells. Monoclonal antibodies have been used as therapeutic checkpoint inhibitors. Through the gut-anticancer axis, the gut microbiota could activate cytotoxic T cells through undefined mechanisms including the effect of short-chain fatty acids (SCFAs). The arrows indicate stimulation and/or augmentation. Note that some critical events have been omitted for clarity. CTLA4: Cytotoxic T-lymphocyte-associated protein 4; PD1: programmed cell death protein 1; PDL1: programmed cell death ligand 1; CD: cluster of differentiation; ROS: reactive oxygen species.
GUT-ANTICANCER AXIS IN CANCER THERAPY
It is generally recognized that commensal microorganisms may coexist within the human gut and other organs protecting against several cancers. For example, gut microbiota dysbiosis of a cancer patient has been identified as a prognostic factor for the success of cancer treatment[36]. In addition, the role of the microbiota in the effectiveness of anti-tumor therapeutics has been designated in various cancers including colorectal cancer, melanoma, and/or sarcoma[37]. The gut mucosal defense system might employ immunological barriers against cancers. Several cohort studies have revealed that patients with colorectal cancer with a great amount of Fusobacterium nucleatum live considerably shorter than patients with lower counts of this bacteria species[38]. Furthermore, it has been documented that some chemotherapies may play a key role in modifying the fecal microbiota of breast cancer patients[39]. Mice transfaunated with modified microbiota communities have been shown to harbor fewer neoplastic lesions in the colon[40]. Several therapeutics are offered with which to change the composition of gut microbiota by the administration of probiotics and/or prebiotics, which could provide various benefits for the treatment of colorectal cancer[41]. In this way, probiotics have been exploited to control gut microbiota, which are defined as living microorganisms with a great health benefit on the administered individuals[42]. Probiotics could also decrease the side effects of various cancer therapies[43]. In addition, the administration of probiotics has decreased the infection incidence of pneumonia after anti-cancer surgery[44]. Accumulating data reveal that modifications in the gut microbiome may contribute to the development, prognosis, and management of several diseases, including various cancers, via the interaction with the immune system [Figure 1]. For example, abundant single species in the gut mucosa, Akkermansia muciniphila, might contribute to the efficacy of PD1 blockade immunotherapy[45]. Some compounds may possess the beneficial properties of gut bacteria, such as short-chain fatty acids to modulate immune responses[46]. Prebiotics, which could stimulate the growth of certain microorganisms, may also trigger modifications in the composition of gut microbiota deliberating such great health benefits[47]. Furthermore, they may be critical to the pharmacological effects of cancer chemotherapies[48]. Thus, there is evidence highlighting the significance of the connections between commensal bacteria and both cancer and immune cells in controlling the effectiveness of cancer therapy, suggesting that gut bacteria could also affect the response to therapeutic cancer medications[49]. As mentioned above, gut bacteria are closely linked to targeted immunotherapies such as anti-PDL1 and anti-CLTA4 blockade therapies[50]. At the same time, anti-cancer treatments may modify the gut microbiota composition, disturbing host homeostasis[51]. The mechanistic elucidation of gut microbiota should be critical before personalized medicine can be made by modulation of the gut microbiota. More comprehension of how gut microbiota influences cancers and immunity will help us find better therapies to manage and treat malignant disease.
PROBIOTICS AS A THERAPEUTIC INTERMEDIATION AGAINST CANCER
The microbiota protects and/or damages host cells including immune cells, indicating that commensal microbes change host immunity, which might contribute to the homeostasis of the host[52]. Generally, immune cells release ROS, which may cause DNA damage[53]. Therefore, inflammation and oxidative stress are thought to be involved in carcinogenesis, enhancing the further release of ROS[54]. Some key physiological roles of ROS include the regulation of enzymes involved in DNA synthesis and DNA repair[55]. For example, several studies have revealed that nickel-iron could bring DNA damage via the DNA binding and/or ROS production, and nickel-iron could also suppress the function of DNA repair systems containing homologous recombination repair, mismatch repair, nucleotide repair, base excision repair, and nonhomologous repair pathways[56]. Oral nickel consumption definitely alters the interaction between the host and the intestinal microbial flora[57]. ROS are defined as oxygen-containing active molecules capable of reacting with numerous biologically important molecules, which may contain various reactive radicals such as hydroxyl radicals and superoxide anions as well as non-radical species including hydrogen peroxide mostly derived from intracellular metabolism and/or inflammatory reactions[58]. One of the leading intracellular resources of ROS is the electron transport chain reaction in mitochondrial throughout ATP synthesis[59]. ROS were initially believed to function as molecules to damage various components in a cell. Now, many studies have established that ROS also play a crucial role in various cellular processes[60]. Certain levels of ROS affect the signaling pathway to regulate mRNA and/or protein expression, which determines the cell destiny, thus profoundly modulating either cell survival or cell apoptosis[61]. Cancer treatments designed to produce excessive concentrations of ROS have been developed to promote cancer cell death and/or apoptosis through considerable oxidative damages, which may include plasma membrane composition damage of cancer cells via the oxidation of both lipid bilayer and membrane proteins[62]. ROS may also influence the ability of dendritic cells (DCs) such as CD8-positive T cells[63]. DCs are known as chief players in coordinating the immune responses, and their interaction with intestinal microbiota is critical for gut homeostasis[64]. The DCs from older animals indicate signs of mitochondrial dysfunction, suggesting an increased ROS production compared to DCs from younger animals[65]. This finding indicates that an intensification in the environmental redox could impede DC function, although DCs actively utilize ROS to enable their important functions.
Probiotics are a group of bacteria that play a crucial role in gut microbiota homeostasis. They are recognized to be valuable in preventing various diseases including inflammatory bowel disease, which is related to ROS levels in animal models[66]. Certain gut bacteria are linked to greater levels of ROS induced by higher fat intake[67]. Microbes that can exert a protective effect at the gastrointestinal level by preventing host DNA damage are considered important probiotics for reducing gut pathologies[68]. Recently, it has been suggested that probiotics might possess a potential molecular mechanism through the action of miRNAs. For example, the useful properties of probiotics could be elucidated based on the pivotal role of certain miRNAs[69], which is addressed in the next section. Owing to the amplified expression of tumor suppressors and/or reduced level of oncogenes after treatment with probiotics, they may be considered valuable for cancer therapy[70]. Future treatments against cancers will likely comprise strategies for modifying the environment at distinct locations in a bearable manner. Therefore, understanding redox regulation of various physiological processes is imperative for evolving new therapeutic approaches.
RELATIONSHIP AMONG GUT MICROBIOTA, ROS, POST-TRANSCRIPTIONAL REGULATION, AND APRO PROTEINS IN CARCINOGENESIS
B-cell translocation gene 1 (BTG1) is a participant of the anti-proliferative (APRO) family proteins, which prevent cancer cell growth and stimulate cancer cell apoptosis[71]. Interestingly, the expression of BTG1 in gut intraepithelial lymphocytes is high[72]. Overexpression of BTG1 along with X-ray irradiation might induce the production of ROS, enhancing the radiation sensitivity of human breast cancer to therapy[73]. Radiotherapy is one of the most effective strategies for breast cancer therapeutics[74]. BTG1 overexpression is involved in the inhibition of PI3K/AKT signaling pathways, which induce the growth inhibition of cancer cells[73]. Generation of ROS is also the common event mediating BTG2[75], which has been revealed to make cancer cells more sensitive to anti-solid tumor treatment with doxorubicin by upregulating SOD2 expression[76]. In response to DNA damages, transducer of ErbB 1 (TOB1), another member of the APRO family, increases due to transcriptional and/or protein stabilization mechanisms, resulting in inhibition of apoptotic signaling[77]. In this way, ROS-mediated cell responses, including several carcinogenesis ones, appear to be involved in the functions of the APRO family. Actually, the expression of an APRO protein is often downregulated in various cancer cells[78]. The APRO genes are categorized as immediate early responsive genes[79]. Gene products of the APRO family include BTG1, pheochromocytoma cell 3 (PC3)/ tetradecanoyl phorbol acetate-inducible sequences 21/BTG2, Abundant in Neuroepithelium Area /BTG3, TOB1, TOB2, PC3B, and others[79], which have been shown to contribute to diverse cellular activities including DNA repair, mRNA stability, and cell-division[80,81].
APRO proteins have been shown to have diverse and important roles; therefore, the molecular mechanisms competent in the proteins may be multifactorial. Some members of the APRO family have been revealed to be involved in the regulation of mRNA transcription by mRNA deadenylation in nucleus and cytoplasm[82]. The N-terminal well-preserved APRO domain has been revealed as the segment for protein-protein interaction, which is able to associate with DNA-binding transcription factors as well as with the deadenylase subunits (CNOT7 and CNOT8) of the carbon catabolite repression 4-negative on TATA-less protein complex[83]. For example, TOB1 can connect to the poly-A nuclease complex CCR4- Chromatin assembly factor-1 (CAF1) and/or the cytoplasmic poly-A binding protein (PABP), indicating the roles of TOB1 in an assembly of mRNA deadenylation machinery[84]. It has also been shown that both TOB1 and TOB2 proteins possess a conserved C-terminal domain with two PABP-interacting motif 2[85]. These TOB proteins can associate with CAF1 and PABP at the same time, which can induce the deadenylation of mRNAs[86]. Remarkably, the anti-proliferative properties of TOB1 have been proposed to depend on the association to the CAF1-CCR4 deadenylase complex[86], indicating that TOB proteins can employ their anti-proliferative activity by modifying mRNA turnover [Figure 2]. BTG2 also binds to the CAF1 deadenylase via the APRO domain for regulating cell proliferation[87]. It has been revealed that mRNA destabilization by the roles of BTG1 and/or BTG2 could retain the cell quiescence[88]. miRNAs could prevent mRNAs function by pairing to the 3’ UTR of the target mRNAs, which later prevents mRNA translation by introducing poly-A tail deadenylation and mRNA destabilization [Figure 2][89]. miRNA-mediated specific deadenylation is reliant on the activity of CAF1 and PABP, which act as miRNA-related coactivators[90]. mRNA deadenylation may also occur following the main translational inhibition, indicating an action of miRNA on mRNA destabilization[90,91]. The miRNA burdened RNA-induced silencing complex (miRISC) might interact with the PABP, CAF1, and CCR4 deadenylases[90,91]. Remarkably, a central module of miRISC might associate with PABP through its C-terminal region, which is essential for miRNA-mediated deadenylation[90,91]. Since APRO proteins could interact with the CAF1-CCR4 complex, APRO proteins might be a modulator of the roles of miRNA [Figure 2]. In addition, the CAF1-CCR4 complex has been revealed as a multifunctional controller that might play important roles in the cellular processes of eukaryotes[92]. Based on the various roles of APRO proteins, the expressions might be regulated by many miRNAs. Actually, miR-21 has been revealed to control BTG2 gene expression during carcinogenesis[93]. More understanding of miRNA could help to improve cancer diagnosis and/or therapy.
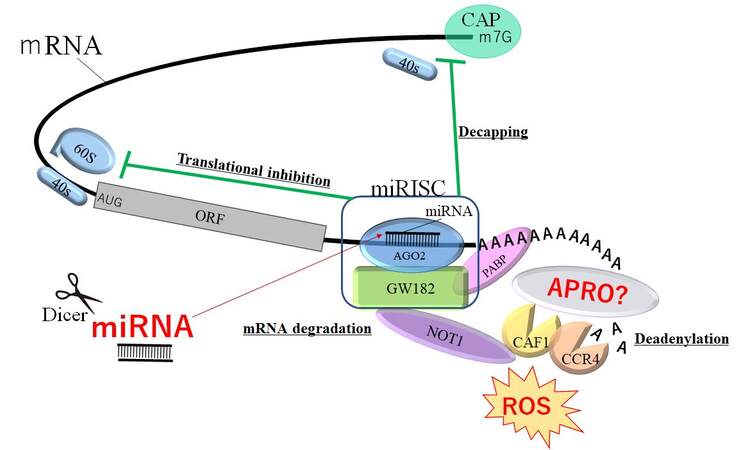
Figure 2. Schematic demonstration of miRNA-mediated inhibition of mRNAs. The GW182 protein interacts with AGO2 protein assembling the miRISC complex, which may enable the deadenylation and mRNAs degradation by PABP and APRO proteins with the CAF1-CCR4-NOT1 complex. The CAF1-CCR4-NOT1 complex is recruited to the 3’ UTR of specific mRNAs through interaction with PABP protein. APRO proteins may also associate with the PABP protein and recruit the CAF1-CCR4-NOT1 complex. Consequently, miRNAs could play dynamic roles in regulating mRNA expression via the decapping, translational inhibition, deadenylation, and degradation of mRNAs. The hammerhead represents inhibition. Note that some critical pathways have been omitted for clarity. AGO2: Argonaute2; PABP: poly(A)-binding protein; APRO: anti-proliferative; ORF: open reading frame; ROS: reactive oxygen species; miRISC: microRNA-induced silencing complex; AUG: initiating codon ATG; CAF1: chromatin assembly factor-1; CCR4-NOT: carbon catabolite repression 4-negative on TATA-less.
FUTURE PERSPECTIVES
Broad studies have provided evidence on the involvement of the gut microbiota in regulating the pathways related to carcinogenesis[94]. In particular, gut dysbiosis has been persuasively linked to some processes of carcinogenesis, which could be oncogenic over the progression of gut mucosal inflammation[95,96]. It is, therefore, also possible that microorganisms adjacent to and/or distant from tumors might disturb cancer initiation, progression, and the course of the disease. Now, gut microbiota is an emergent target in the next cancer therapeutics. Currently, there are some therapeutic methods accessible that can change gut microbiota[97]. In particular, the administration of prebiotics and probiotics seems to be beneficial in certain circumstances[97]. Additionally, crucial studies have established the significant impact of fecal microbiota transplantation on the host immune responses[98]. Administration of certain probiotic bacteria appears to support cancer treatment owing to their immunomodulatory activities and the ability to decrease adverse effects of anti-cancer therapy[99]. In particular, the uncomfortable events after chemotherapy as well as radiotherapy could be decreased by probiotic administration[100]. Furthermore, we found that the TOB1 expression levels in colon epithelium have been increased and/or upregulated by stimulation with certain microorganisms (unpublished). If so, gut microbiota could potentiate the effect of cancer therapy. Consistently, the growth suppression by adenovirus-mediated TOB1 protein in pancreatic cancer has been described, suggesting an application of APRO proteins for chemotherapy-resistant cancerous peritonitis[101]. Identification of such supportive mechanisms may provide a promising viewpoint for microbiota-based therapies, which could be integrated with general cancer therapy to improve the outcomes of patients [Figure 3]. However, this requires intensive research in the future. Future analyses are also mandatory to elucidate the mechanisms by which the microbiome may assist as a basis for diagnostics and/or treatments for cancer. Moreover, there is a strong necessity to assess the safety of probiotics and/or fecal microbiota transplantation with regard to their long-term clinical effects.
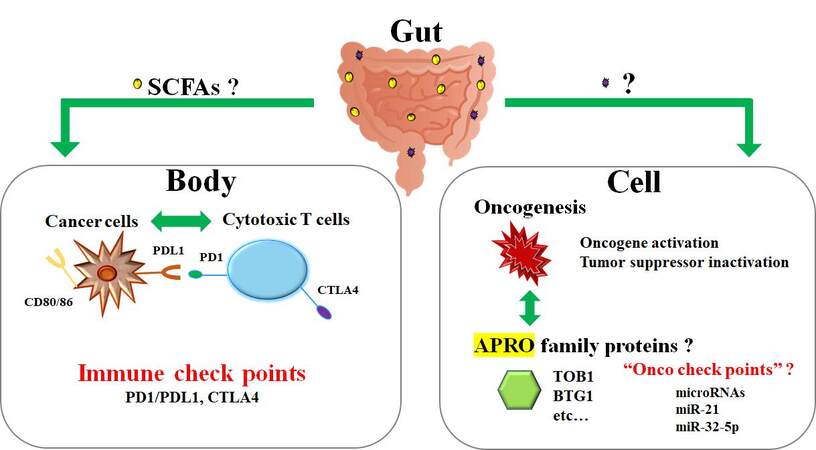
Figure 3. The relationship between cytotoxic T cells and cancer cells in a body resembles the relationship between tumor suppressor APRO family proteins and oncogenesis in a cell. It could be hypothesized that the former is regulated with immune checkpoint molecules such as PD1/PDL1 and CTLA4, whereas the latter may be regulated by various miRNAs. The gut commensal microbiota might support both phases of the anti-cancer activity via unclear factors, possibly including ROS and/or SCFAs. APRO: Anti-proliferative; CTLA4: cytotoxic T-lymphocyte-associated protein 4; PD1: programmed cell death protein 1; PDL1: programmed cell death ligand 1; BTG1: B-cell translocation gene 1; TOB: transducer of ErbB-2; ROS: reactive oxygen species; SCFAs: short-chain fatty acids.
DECLARATIONS
Authors’ contributionsSawamura H and Matsuda S contributed to conception of the study.
Sawamura H, Ikeda Y, Taniguchi K, Tsuji A, Kitagishi Y and Matsuda S wrote sections of the manuscript.
All authors contributed to manuscript revision, read, and had given final approval.
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestAll authors declared that they have no competing financial interests.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2022.
REFERENCES
1. Paray BA, Albeshr MF, Jan AT, Rather IA. Leaky gut and autoimmunity: an intricate balance in individuals health and the diseased state. Int J Mol Sci 2020;21:9770.
2. Laursen MF. Gut Microbiota Development: influence of diet from infancy to toddlerhood. Ann Nutr Metab 2021:1-14.
3. Reedy AR, Luo L, Neish AS, Jones RM. Commensal microbiota-induced redox signaling activates proliferative signals in the intestinal stem cell microenvironment. Development 2019;146:dev171520.
4. Campbell EL, Colgan SP. Control and dysregulation of redox signalling in the gastrointestinal tract. Nat Rev Gastroenterol Hepatol 2019;16:106-20.
5. Lucas K, Morris G, Anderson G, Maes M. The Toll-Like receptor radical cycle pathway: a new drug target in immune-related chronic fatigue. CNS Neurol Disord Drug Targets 2015;14:838-54.
6. Saint-Georges-Chaumet Y, Attaf D, Pelletier E, Edeas M. Targeting microbiota-mitochondria inter-talk: microbiota control mitochondria metabolism. Cell Mol Biol (Noisy-le-grand) 2015;61:121-4.
7. Liu S, Hu J, Shi C, et al. Sparstolonin B exerts beneficial effects on prostate cancer by acting on the reactive oxygen species-mediated PI3K/AKT pathway. J Cell Mol Med 2021;25:5511-24.
8. Zhao Y, Ma T, Zhang Z, et al. Resolvin D1 attenuates acid-induced DNA damage in esophageal epithelial cells and rat models of acid reflux. Eur J Pharmacol 2021;912:174571.
9. Zhang Z, Wang Y, Li Q. Mechanisms underlying the effects of stress on tumorigenesis and metastasis (Review). Int J Oncol 2018;53:2332-42.
10. Falone S, Santini S Jr, Cordone V, et al. Extremely low-frequency magnetic fields and redox-responsive pathways linked to cancer drug resistance: insights from co-exposure-based. in vitro ;6:33.
11. Szczyrek M, Mlak R, Krawczyk P, et al. Polymorphisms of genes encoding multidrug resistance proteins as a predictive factor for second-line docetaxel therapy in advanced non-small cell lung cancer. Pathol Oncol Res 2017;23:607-14.
12. Iranmanesh Y, Jiang B, Favour OC, et al. Mitochondria’s role in the maintenance of cancer stem cells in glioblastoma. Front Oncol 2021;11:582694.
13. Keenan J, Murphy L, Henry M, Meleady P, Clynes M. Proteomic analysis of multidrug-resistance mechanisms in adriamycin-resistant variants of DLKP, a squamous lung cancer cell line. Proteomics 2009;9:1556-66.
14. Kang YB, Cai Y. Faecal microbiota transplantation enhances efficacy of immune checkpoint inhibitors therapy against cancer. World J Gastroenterol 2021;27:5362-75.
15. Grenda A, Krawczyk P. Cancer trigger or remedy: two faces of the human microbiome. Appl Microbiol Biotechnol 2021;105:1395-405.
16. Encarnação JC, Pires AS, Amaral RA, et al. Butyrate, a dietary fiber derivative that improves irinotecan effect in colon cancer cells. J Nutr Biochem 2018;56:183-92.
17. Santo C, Caseiro C, Martins MJ, Monteiro R, Brandão I. Gut microbiota, in the halfway between nutrition and lung function. Nutrients 2021;13:1716.
19. Sultan S, El-Mowafy M, Elgaml A, et al. Metabolic influences of gut microbiota dysbiosis on inflammatory bowel disease. Front Physiol 2021;12:715506.
20. Chen Y, Chen YX. Microbiota-associated metabolites and related immunoregulation in colorectal cancer. Cancers (Basel) 2021;13:4054.
21. Jalandra R, Dalal N, Yadav AK, et al. Emerging role of trimethylamine-N-oxide (TMAO) in colorectal cancer. Appl Microbiol Biotechnol 2021;105:7651-60.
22. Grover K, Gregory S, Gibbs JF, Emenaker NJ. A discussion of the gut microbiome’s development, determinants, and dysbiosis in cancers of the esophagus and stomach. J Gastrointest Oncol 2021;12:S290-300.
23. Temraz S, Nassar F, Kreidieh F, et al. Hepatocellular carcinoma immunotherapy and the potential influence of gut microbiome. Int J Mol Sci 2021;22:7800.
24. Brandi G, De Lorenzo S, Candela M, et al. Microbiota, NASH, HCC and the potential role of probiotics. Carcinogenesis 2017;38:231-40.
25. Golombos DM, Ayangbesan A, O’Malley P, et al. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology 2018;111:122-8.
26. Smith KS, Frugé AD, van der Pol W, et al. Gut microbial differences in breast and prostate cancer cases from two randomised controlled trials compared to matched cancer-free controls. Benef Microbes 2021;12:239-48.
27. Alpuim Costa D, Nobre JG, Batista MV, et al. Human microbiota and breast cancer-is there any relevant link? Front Microbiol 2021;12:584332.
28. Gadelle D, Raibaud P, Sacquet E. Beta-Glucuronidase activities of intestinal bacteria determined both. in vitro ;49:682-5.
29. Liu X, Cheng Y, Zang D, et al. The role of gut microbiota in lung cancer: from carcinogenesis to immunotherapy. Front Oncol 2021;11:720842.
30. Shi Z, Zhang M. Emerging roles for the gut microbiome in lymphoid neoplasms. Clin Med Insights Oncol 2021;15:11795549211024197.
31. Mitsuhashi K, Nosho K, Sukawa Y, et al. Association of fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 2015;6:7209-20.
32. Baysal B, İnce AT, Gültepe B, et al. Helicobacter pylori is undetectable in intraductal papillary mucinous neoplasm. Pancreatology 2016;16:865-8.
33. Rabelo-Gonçalves EM, Roesler BM, Zeitune JM. Extragastric manifestations of Helicobacter pylori infection: Possible role of bacterium in liver and pancreas diseases. World J Hepatol 2015;7:2968-79.
34. Ketpueak T, Thiennimitr P, Apaijai N, Chattipakorn SC, Chattipakorn N. Association of chronic opisthorchis infestation and microbiota alteration on tumorigenesis in cholangiocarcinoma. Clin Transl Gastroenterol 2020;12:e00292.
35. Jauvain M, Courtois S, Lehours P, Bessède E. Metformin modifies the gut microbiota of mice infected with. helicobacter pylori ;14:329.
36. Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science 2018;359:1366-70.
37. Veziant J, Villéger R, Barnich N, Bonnet M. Gut microbiota as potential biomarker and/or therapeutic target to improve the management of cancer: focus on colibactin-producing. escherichia coli ;13:2215.
38. van Praagh JB, de Goffau MC, Bakker IS, et al. Mucus microbiome of anastomotic tissue during surgery has predictive value for colorectal anastomotic leakage. Ann Surg 2019;269:911-6.
39. Zidi O, Souai N, Raies H, et al. Fecal metabolic profiling of breast cancer patients during neoadjuvant chemotherapy reveals potential biomarkers. Molecules 2021;26:2266.
40. Parker KD, Maurya AK, Ibrahim H, et al. Dietary rice bran-modified human gut microbial consortia confers protection against colon carcinogenesis following fecal transfaunation. Biomedicines 2021;9:144.
41. Kaźmierczak-Siedlecka K, Daca A, Fic M, et al. Therapeutic methods of gut microbiota modification in colorectal cancer management - fecal microbiota transplantation, prebiotics, probiotics, and synbiotics. Gut Microbes 2020;11:1518-30.
42. Mehmood K, Moin A, Hussain T, et al. Can manipulation of gut microbiota really be transformed into an intervention strategy for cardiovascular disease management? Folia Microbiol (Praha) 2021;66:897-916.
43. Waziri A, Bharti C, Aslam M, et al. Probiotics for the chemoprotective role against the toxic effect of cancer chemotherapy. Anticancer Agents Med Chem 2022;22:654-67.
44. Liu ZH, Huang MJ, Zhang XW, et al. The effects of perioperative probiotic treatment on serum zonulin concentration and subsequent postoperative infectious complications after colorectal cancer surgery: a double-center and double-blind randomized clinical trial. Am J Clin Nutr 2013;97:117-26.
45. Wang Y, Ma R, Liu F, Lee SA, Zhang L. Modulation of gut microbiota: a novel paradigm of enhancing the efficacy of programmed death-1 and programmed death ligand-1 blockade therapy. Front Immunol 2018;9:374.
46. Mo Q, Liu T, Fu A, et al. Novel gut microbiota patterns involved in the attenuation of dextran sodium sulfate-induced mouse colitis mediated by glycerol monolaurate via inducing anti-inflammatory responses. mBio 2021;12:e0214821.
47. Blanco-Pérez F, Steigerwald H, Schülke S, et al. The dietary fiber pectin: health benefits and potential for the treatment of allergies by modulation of gut microbiota. Curr Allergy Asthma Rep 2021;21:43.
49. Panebianco C, Andriulli A, Pazienza V. Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome 2018;6:92.
50. Alexander JL, Wilson ID, Teare J, et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol 2017;14:356-65.
51. Gagnière J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol 2016;22:501-18.
52. Pellon A, Barriales D, Peña-Cearra A, et al. The commensal bacterium. Lactiplantibacillus plantarum ;13:1939598.
53. Zhu W, Liu Y, Zhang W, et al. Selenomethionine protects hematopoietic stem/progenitor cells against cobalt nanoparticles by stimulating antioxidant actions and DNA repair functions. Aging (Albany NY) 2021;13:11705-26.
54. Genua F, Raghunathan V, Jenab M, Gallagher WM, Hughes DJ. The role of gut barrier dysfunction and microbiome dysbiosis in colorectal cancer development. Front Oncol 2021;11:626349.
55. Halcrow PW, Lynch ML, Geiger JD, Ohm JE. Role of endolysosome function in iron metabolism and brain carcinogenesis. Semin Cancer Biol 2021;76:74-85.
56. Guo H, Liu H, Wu H, et al. Nickel carcinogenesis mechanism: DNA damage. Int J Mol Sci 2019;20:4690.
57. Zhou X, Li J, Sun JL. Oral nickel changes of intestinal microflora in mice. Curr Microbiol 2019;76:590-6.
58. Borsani B, De Santis R, Perico V, et al. The role of carrageenan in inflammatory bowel diseases and allergic reactions: where do we stand? Nutrients 2021;13:3402.
59. Selivanov VA, Zagubnaya OA, Nartsissov YR, Cascante M. Unveiling a key role of oxaloacetate-glutamate interaction in regulation of respiration and ROS generation in nonsynaptic brain mitochondria using a kinetic model. PLoS One 2021;16:e0255164.
60. Canton M, Sánchez-Rodríguez R, Spera I, et al. Reactive oxygen species in macrophages: sources and targets. Front Immunol 2021;12:734229.
61. Meijles DN, Zoumpoulidou G, Markou T, et al. The cardiomyocyte “redox rheostat”: Redox signalling via the AMPK-mTOR axis and regulation of gene and protein expression balancing survival and death. J Mol Cell Cardiol 2019;129:118-29.
62. Davoodi P, Srinivasan MP, Wang CH. Effective co-delivery of nutlin-3a and p53 genes via core-shell microparticles for disruption of MDM2-p53 interaction and reactivation of p53 in hepatocellular carcinoma. J Mater Chem B 2017;5:5816-34.
63. Choi JA, Cho SN, Lee J, et al. Lipocalin 2 regulates expression of MHC class I molecules in mycobacterium tuberculosis-infected dendritic cells via ROS production. Cell Biosci 2021;11:175.
64. Moreira TG, Mangani D, Cox LM, et al. PD-L1+ and XCR1+ dendritic cells are region-specific regulators of gut homeostasis. Nat Commun 2021;12:4907.
65. Chougnet CA, Thacker RI, Shehata HM, et al. Loss of phagocytic and antigen cross-presenting capacity in aging dendritic cells is associated with mitochondrial dysfunction. J Immunol 2015;195:2624-32.
66. Din AU, Hassan A, Zhu Y, et al. Inhibitory effect of bifidobacterium bifidum ATCC 29521 on colitis and its mechanism. J Nutr Biochem 2020;79:108353.
67. Lee JY, Bae E, Kim HY, et al. High-fat-diet-induced oxidative stress linked to the increased colonization of lactobacillus. sakei ;9:e0007421.
68. Garcia-Gonzalez N, Prete R, Perugini M, et al. Probiotic antigenotoxic activity as a DNA bioprotective tool: a minireview with focus on endocrine disruptors. FEMS Microbiol Lett 2020;367:fnaa041.
69. Davoodvandi A, Marzban H, Goleij P, et al. Effects of therapeutic probiotics on modulation of microRNAs. Cell Commun Signal 2021;19:4.
70. Heydari Z, Rahaie M, Alizadeh AM, et al. Effects of lactobacillus acidophilus and bifidobacterium bifidum probiotics on the expression of microRNAs 135b, 26b, 18a and 155, and their involving genes in mice colon cancer. Probiotics Antimicrob Proteins 2019;11:1155-62.
71. Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell 2014;26:262-72.
72. Shires J, Theodoridis E, Hayday AC. Biological insights into TCRγδ+ and TCRαβ+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE). Immunity 2001;15:419-34.
73. Zhu R, Li W, Xu Y, Wan J, Zhang Z. Upregulation of BTG1 enhances the radiation sensitivity of human breast cancer in vitro and in vivo. Oncol Rep 2015;34:3017-24.
75. Imran M, Lim IK. Regulation of Btg2(/TIS21/PC3) expression via reactive oxygen species-protein kinase C-ΝFκΒ pathway under stress conditions. Cell Signal 2013;25:2400-12.
76. Sundaramoorthy S, Ryu MS, Lim IK. B-cell translocation gene 2 mediates crosstalk between PI3K/Akt1 and NFκB pathways which enhances transcription of MnSOD by accelerating IκBα degradation in normal and cancer cells. Cell Commun Signal 2013;11:69.
77. Suzuki T, Tsuzuku J, Hayashi A, et al. Inhibition of DNA damage-induced apoptosis through Cdc7-mediated stabilization of Tob. J Biol Chem 2012;287:40256-65.
78. Guan R, Peng L, Wang D, et al. Decreased TOB1 expression and increased phosphorylation of nuclear TOB1 promotes gastric cancer. Oncotarget 2017;8:75243-53.
79. Matsuda S, Rouault J, Magaud J, Berthet C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Letters 2001;497:67-72.
80. Doidge R, Mittal S, Aslam A, Winkler GS. The anti-proliferative activity of BTG/TOB proteins is mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the Ccr4-not complex. PLoS One 2012;7:e51331.
81. Yang X, Morita M, Wang H, et al. Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic Acids Res 2008;36:6872-81.
82. Amine H, Ripin N, Sharma S, et al. A conserved motif in human BTG1 and BTG2 proteins mediates interaction with the poly(A) binding protein PABPC1 to stimulate mRNA deadenylation. RNA Biol 2021;18:2450-65.
83. Chapat C, Chettab K, Simonet P, et al. Alternative splicing of CNOT7 diversifies CCR4-NOT functions. Nucleic Acids Res 2017;45:8508-23.
84. Horiuchi M, Takeuchi K, Noda N, et al. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J Biol Chem 2009;284:13244-55.
85. Lim NS, Kozlov G, Chang TC, et al. Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein. Implications for HYD function. J Biol Chem 2006;281:14376-82.
86. Ezzeddine N, Chen CY, Shyu AB. Evidence providing new insights into TOB-promoted deadenylation and supporting a link between TOB’s deadenylation-enhancing and antiproliferative activities. Mol Cell Biol 2012;32:1089-98.
87. Stupfler B, Birck C, Séraphin B, Mauxion F. BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat Commun 2016;7:10811.
88. Hwang SS, Lim J, Yu Z, et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 2020;367:1255-60.
89. Räsch F, Weber R, Izaurralde E, Igreja C. 4E-T-bound mRNAs are stored in a silenced and deadenylated form. Genes Dev 2020;34:847-60.
90. Fabian MR, Mathonnet G, Sundermeier T, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 2009;35:868-80.
91. Flamand MN, Wu E, Vashisht A, et al. Poly(A)-binding proteins are required for microRNA-mediated silencing and to promote target deadenylation in C. elegans. Nucleic Acids Res 2016;44:5924-35.
92. Collart MA, Panasenko OO. The Ccr4-Not Complex: Architecture and Structural Insights. Subcell Biochem 2017;83:349-79.
93. Mao B, Zhang Z, Wang G. BTG2: a rising star of tumor suppressors (review). Int J Oncol 2015;46:459-64.
94. Peng C, Ouyang Y, Lu N, Li N. The NF-κB Signaling Pathway, the Microbiota, and Gastrointestinal Tumorigenesis: Recent Advances. Front Immunol 2020;11:1387.
95. Gu S, Zaidi S, Hassan MI, et al. Mutated ceacams disrupt transforming growth factor beta signaling and alter the intestinal microbiome to promote colorectal carcinogenesis. Gastroenterology 2020;158:238-52.
96. Lang M, Baumgartner M, Rożalska A, et al. Crypt residing bacteria and proximal colonic carcinogenesis in a mouse model of lynch syndrome. Int J Cancer 2020;147:2316-26.
97. Ke A, Parreira VR, Goodridge L, Farber JM. Current and future perspectives on the role of probiotics, prebiotics, and synbiotics in controlling pathogenic. cronobacter ;12:755083.
98. Genton L, Lazarevic V, Stojanovic O, et al. Metataxonomic and metabolic impact of fecal microbiota transplantation from patients with pancreatic cancer into germ-free mice: a pilot study. Front Cell Infect Microbiol 2021;11:752889.
99. Sambrani R, Abdolalizadeh J, Kohan L, Jafari B. Recent advances in the application of probiotic yeasts, particularly saccharomyces, as an adjuvant therapy in the management of cancer with focus on colorectal cancer. Mol Biol Rep 2021;48:951-60.
100. Lima WG, Pessoa RM, Vital KD, et al. Effect of probiotics on the maintenance of intestinal homeostasis after chemotherapy: systematic review and meta-analysis of pre-clinical studies. Benef Microbes 2020;11:305-18.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Sawamura H, Taniguchi K, Ikeda Y, Tsuji A, Kitagishi Y, Matsuda S. Roles of gut dysbiosis, anti-proliferative proteins, and post-transcriptional regulation in carcinogenesis. J Transl Genet Genom 2022;6:157-68. http://dx.doi.org/10.20517/jtgg.2021.57
AMA Style
Sawamura H, Taniguchi K, Ikeda Y, Tsuji A, Kitagishi Y, Matsuda S. Roles of gut dysbiosis, anti-proliferative proteins, and post-transcriptional regulation in carcinogenesis. Journal of Translational Genetics and Genomics. 2022; 6(2): 157-68. http://dx.doi.org/10.20517/jtgg.2021.57
Chicago/Turabian Style
Sawamura, Haruka, Kurumi Taniguchi, Yuka Ikeda, Ai Tsuji, Yasuko Kitagishi, Satoru Matsuda. 2022. "Roles of gut dysbiosis, anti-proliferative proteins, and post-transcriptional regulation in carcinogenesis" Journal of Translational Genetics and Genomics. 6, no.2: 157-68. http://dx.doi.org/10.20517/jtgg.2021.57
ACS Style
Sawamura, H.; Taniguchi K.; Ikeda Y.; Tsuji A.; Kitagishi Y.; Matsuda S. Roles of gut dysbiosis, anti-proliferative proteins, and post-transcriptional regulation in carcinogenesis. J. Transl. Genet. Genom. 2022, 6, 157-68. http://dx.doi.org/10.20517/jtgg.2021.57
About This Article
Copyright
Data & Comments
Data
 Cite This Article 11 clicks
Cite This Article 11 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.