Large-scale genomic data-mining implicates dysregulated nuclear receptor-mediated signaling in mental illness
Abstract
Aim: Mental illness comprises a group of heterogeneous conditions attributable to a complex interplay between hereditary and environmental components. Acting at the interface between environmental stimuli and their genomic actions, nuclear receptors (NRs) appear uniquely suited to facilitate gene-environment interactions in the context of mental health. Genetic disruptions to the NR transcriptomic complex (NTC) give rise to neuropsychiatric pathologies, and epidemiological risks involving a steroid response are among the most replicated in psychiatry. Importantly, pharmacological targeting of NR-mediated signaling is clinically effective in the treatment of psychiatric disorders. Here, we systematically interrogated large-scale deposited data to provide a comprehensive evaluation of the genomic NTC risk burden in mental illness.
Methods: Utilizing data from large, recent genome-, exome-, and methylome-wide association studies on psychiatric disorders, we assessed the representation of NTC genes among top associated loci and tested the gene set associations for NTC and NR target genes using GWAS summary statistics. Through data mining and transcriptomic profiling of NR-mediated signaling in the diseased and healthy human brain, we categorized psychiatry-relevant NTC gene networks.
Results: We found that NTC genes are significantly overrepresented in genome-, methylome-, and exome-wide associated loci and that the NTC, as well as NR target gene sets, is overall associated with mental illness. Accordingly, we identified transcriptomic NTC signatures in patient brain samples. In line with a key role for orchestrated NR-mediated signaling in the developing brain, particularly NTC co-expression networks with prenatal peak expression are enriched with differentially methylated, sex-biased, and psychiatry-associated risk variants.
Conclusion: Here, we provide multilevel evidence that supports genomic NR-mediated signaling as a common and core molecular mechanism in mental illness, and we highlight specific NR-signaling pathways with putative diagnostic and pharmacological intervention potential in psychiatry.
Keywords
INTRODUCTION
Psychiatric disorders (PDs) comprise a heterogeneous group of conditions collectively characterized by changes in patterns of thoughts, emotions, and behaviors. Suggestive of interconnected etiologies, clinical and therapeutic profiles are overlapping and identified risks are typically non-specifically associated with a range of mental disorders[1-5]. Most PDs are highly heritable and thousands of genetic variants are likely to contribute[6-11]. The effect of genetic risk is further conditional on environmental factors, resulting in complex gene-environment interactions (GxE)[12]. Understanding how hereditary risk and environmental exposures collectively shape the developing brain and mind is thus key to comprehending the pathobiology of mental illness and the implementation of precision medicine in psychiatry.
Acting at the interface among environmental stimuli, endocrine signaling, and their genomic actions, a group of ligand-inducible transcription factors, nuclear receptors (NRs), appear uniquely suited to facilitate GxE in the context of mental health[13,14]. NRs function as biological sensors that respond to a variety of xenobiotics, steroids, and endogenous lipid- and cholesterol-derived compounds[15,16]. Epidemiological risk factors involving a steroid or steroid-like response are among the most replicated in psychiatry[17-27], and several NR ligands have been associated with PDs (e.g., retinoic acid[28-30], vitamin D[17,19]; stress[31], sex[32-35], and thyroid hormones[36]; endocannabinoids[37,38]; and polyunsaturated fatty acids[39,40]). Upon activation, NRs facilitate fine-tuned transcriptional regulation of defined sets of promotor hormone response element (HRE)-containing target genes in a cell-, tissue-, and developmental-specific manner. In this way, NRs play essential roles in the developing and mature central nervous system (CNS)[41,42] and have crucial and diverse functions in many aspects of human metabolism, reproduction, inflammation, and physiology[41]. Consequently, NRs are highly intolerant to loss of function (LoF) mutations[43], and genetic defects in at least 20 of the 48 NRs encoded by the human genome are associated with pathological states, including neurological disorders and mental illness[41,44]. The latter is highlighted by the severe intellectual disability displayed by autism spectrum disorder (ASD) and epilepsy cases harboring LoF mutations in genes encoding retinoic acid receptor-related orphan receptors (RORA[45] and RORB[46]). Genetic variation in and around a large fraction of NRs has furthermore been associated with PDs and psychiatry-related traits (see Supplementary Table 1 for a summary). The transcriptional activity and specificity of NRs is ensured through a dynamic interplay with a comprehensive, but loosely defined, co-regulator complexome, encompassing > 500 NR coregulators[47,48] - collectively the NR transcriptome complex (NTC). The specific interactions between individual NRs and their coregulators are in part determined by the biophysical binding to NR interaction domains (NRIDs) on the regulators[49]. NR coregulators often contain multiple NRIDs and display overlap in their specificity and affinity for NRs[49]. In addition, genes may contain several different HREs, and NR coregulators may dictate opposite transcriptional outcomes, depending on cellular context[50,51]. The modes by which NR coregulators affect NR action are diverse and include direct recruitment of transcriptional machinery as well as chromatin remodeling, histone modifications, and chaperone activity[52,53]. The complexity of NR coregulator interactions is reflected in the palette of pathologies associated with genetic variation to this group of transcriptional regulators[54]. As has been reported for NRs, LoF mutations in several NR coregulators lead to intellectual disability and mental health problems[55-60]. Supporting an overall increased genetic risk load in NR transcriptional networks in mental illness, genetic variation in loci harboring NR coregulators has been reported in a range of PDs (see Supplementary Table 1 for a summary), and increased polygenic burden in retinoid and glucocorticoid biogenesis and signaling pathways has recently been associated with schizophrenia (SZ) and depression, respectively[18,61]. The importance of NR coregulator-mediated modulation of NR action has further been demonstrated by molecular genetic studies in preclinical models[41], where genetic disruption to NR coregulators generally results in behavioral impairments and neurobiological alterations with translational relevance to PDs[55,62-69]. Collectively, ample evidence implicates dysregulated NR-mediated signaling in the pathoetiology of mental illness, and it is thus conceivable that genetic vulnerability to NR-mediated signaling, in combination with their ligand-associated risk factors, collectively shapes the risk and clinical manifestation of PDs.
Here, we provide a comprehensive and systematic data-mining effort and functional genomic analysis of the NTC in large-scale genetic and epigenetic data and present new evidence that supports dysregulated NR-mediated signaling as a common and core molecular pathway in mental illness with significant diagnostic and therapeutic potential in psychiatry.
METHODS
Gene set selection, filtering, and overlap analyses
NTC gene set
NTC gene set includes genes encoding NRs and NR coregulators in the human genome. A defined list of NR coregulators was obtained by compiling curated entities from the now deprecated Nuclear Receptor Signaling Atlas (NURSA; http://www.nursa.org), NRIDs containing NR coregulators with validated biophysical NR interactions from a recent large-scale peptide array-based study[49], and minimal endogenous modules of NR coregulators identified in a recent comprehensive IP/MS-based study of endogenous human coregulator protein complex networks[48]. The final list consisting of 48 NR encoding genes and 522 NR coregulator-encoding genes can be viewed in Supplementary Table 2.
Genome-wide associated gene sets
For the analysis of overlap between NTC gene sets and genes in genome-wide significant (GWS) loci in PDs, the following PGC/iPSYCH PD GWASs were assessed: SZ[70], bipolar disorder (BPD)[10], major depressive disorder (MDD)[8], ASD[10], attention deficit/hyperactivity disorder (ADHD)[11], and cross-disorder (CD)[4], which includes SZ, BPD, MDD, ASD, ADHD, anorexia nervosa, obsessive-compulsive disorder, and Tourette syndrome. For the illustration of NTC genes (NTCs) among genes in GWS loci in SZ [Figure 1], a smaller PGC GWAS[7] with 108 GWS loci was used with the readability of the illustration in mind. Additionally, the following non-PD GWASs were assessed: Alzheimer’s disease (AD)[71]; type 2 diabetes (T2D)[72], heart failure (HF)[73], body mass index (BMI)[74], height[74], and COVID-19 (positive vs. population) downloaded from GRASP[75] (see Supplementary Table 3 for details). PGC genotype data were all processed using the PGC-developed Ricopili pipeline[76]; thus, to obtain comparable locus boundaries and in turn GWS gene sets, summary statistics from non-PGC studies were similarly processed using Ricopili with 1000 Genomes Project (Phase 3 v5a) as reference.
Figure 1. Genetic support for dysregulated NR-mediated signaling in mental illness. Consistently, ~15% of risk loci in PDs harbor NTC-encoding genes. Manhattan plots show the SNP-based association landscape for each of the five psychiatric disorders [attention deficit hyperactivity disorder (ADHD)[11], schizophrenia (SZ)[7] (for presented analyses, data from a newer, larger GWAS[70] were used), bipolar disorder (BPD), autism spectrum disorder (ASD), and major depressive disorder (MDD)] with the red dotted line marking the significance cut-off for genome-wide significantly associated signals. Brain-expressed protein-coding genes within each locus are shown as columns of tiles, where NTC encoding genes are highlighted in red. GWS: Genome-wide significant; NTC: NR transcriptomic complex; HRE: hormone response element; NR: nuclear receptor.
Exome-wide associated gene sets
For the analysis of overlap between NTC genes and genes harboring rare coding variants (RCVs), only large whole exome sequencing studies (WESs) with > 3000 individuals (patients and healthy controls) identifying genes with RCVs were assessed. This is limited to: SZ[77] and ASD[78] (see Supplementary Table 4 for details).
Methylome-wide associated gene sets
To assess the epigenetic burden on NTC genes in patient blood and the developing fetal brain, we utilized data from large epigenome-wide association studies of common mental disorders, namely SZ[79-81], MDD[82], ADHD[83,84], and ASD[85], as well as a methylomic study of fetal brain development[86]. Although varying between studies, P-value cut-offs were comparable. Looking at the top findings reported by the authors in each study, we removed duplicated gene names and findings that did not map to any gene (for an overview, see Supplementary Table 5).
All gene sets were filtered based on the following criteria: protein-coding and detected (RPKM ≠ 0) in human brain tissue at any developmental stage as assessed in the BrainSpan database[87]. MHC region was excluded from all datasets. For each phenotype, we determined the fraction of protein-coding, brain-expressed genes that overlapped with our list of NTC encoding genes and compared the fractions across studies. Significance of overlap was determined using one-sided chi-squared tests.
Gene set association analyses
Gene set analysis was performed with MAGMA[88] using default settings, based on summary statistics from selected publicly available GWASs (see Supplementary Table 3 for details). SNPs outside protein-coding and brain detected (RPKM ≠ 0) genes, as well as SNPs within the MHC region and imputed SNPs with info score < 0.8, were excluded from the analyses.
For the analysis of promotor HRE containing genes, available curated and non-redundant sets of transcription factor binding sites (TFBSs) for NR monomers, HOCOMOCOv11_core_HUMAN, were downloaded from the HOCOMOCO collection[89] (http://www.cbrc.kaust.edu.sa/hocomoco11) and genomic positions were identified using the FIMO tool (http://meme-suite.org/tools/fimo)[90]. Subsequently, lists were generated for each NR with genes containing their HRE within their promotor sequences (2000 bp upstream of TSS). Gene annotation files contained every human protein encoding gene detected in brain tissue (www.brainspan.org, RPKM ≠ 0 in any sample). For the assessment of similarities between HRE gene sets, pairwise Jaccard similarity coefficients and significance of overlap were calculated using the GeneOverlap R package (version 1.24.0)[91].
Cortical transcriptomic profiling of NTC and HRE gene sets in patients
We used available analyses of differentially expressed genes (DEGs) in the dorsolateral prefrontal cortex of 258 SZ patients and 271 healthy controls from the CommonMind Consortium (CMC; CommonMind.org Synapse ID: syn5607652)[92].
Enrichment analysis of TFBSs was carried out according to Gearing et al.[93] using CiiiDER. Briefly, promotor sequences (2000 bp upstream of TSS) were extracted from the Homo sapiens GRCh38.94 genome file. Identification of TFBSs in these sequences was performed with HOCOMOCOv11_core_HUMAN transcription factor position frequency matrices [downloaded from the HOCOMOCO collection[89]
Brain transcriptomic profile of the nuclear receptor transcriptome complex
Normalized gene expression values (RPKM) for 16 different brain tissues in the developing and mature brain was downloaded from www.brainspan.org[87]. Developmental stages were defined as Prenatal
RESULTS
Common and rare psychiatry-associated genetic variation is enriched with genes implicated in nuclear receptor-mediated signaling
Whereas genetic variation in NR and NR coregulators, individually, has been associated with PDs in association and linkage studies, the genetic risk profile of the NR transcriptome complex (NTC) as a whole has not been systematically assessed at a large-scale, whole-genome level. More than 300 curated NR coregulators have been reported by the Nuclear Receptor Signaling Atlas consortium, but recent efforts have both added to this list and significantly extended the known interactions between NRs and NR coregulators [Supplementary Table 6][48,49]. Hence, we composed a defined list of NTC encoding genes based on curated databases and strictly validated protein-protein interactions (the NR gene set with 48 genes and the NR coregulator gene set with 522 genes) and mapped the overlay of these lists with genes annotated to genome-wide significantly associated (GWS) loci in PDs[7-11,70]. Consistently, ~15% of loci across diagnostic entities harbored NTC encoding genes, except for ASD, where only three GWS loci were identified [Figure 1 and Supplementary Table 7]. In addition, > 13% of all brain-expressed NTC genes reside in loci associated with ADHD, BPD, MDD, SZ, or CD [Figure 1 and Supplementary Table 8]. Individually, this represents a significant overrepresentation in MDD [chi-squared test (one-tailed), P = 0.012] and SZ [chi-squared test (one-tailed), P = 0.002]. Notably, > 20% of NTC genes in GWS loci are associated with two or more PDs (e.g., EP300 and ESR2). A similar overlap was seen for non-psychiatric traits whose biology is closely interlinked with NR-mediated signaling[97-104] [Supplementary Table 7].
Genetic variants displaying GWS account for only the most significant, small fraction of the total heritability of PDs. Hence, to further explore the genetic PD burden in the NTC, we employed a gene set analysis approach based on the aggregated association of individual genetic markers within the NTC gene set[88]. Analyses using the most recently available GWAS summary statistics from each of the five PDs, namely SZ, BPD, ASD, ADHD, and MDD[7-11,70], as well as the currently largest CD GWAS[4], revealed a significant association of the NR gene subset of the NTC to both MDD (P = 0.008) and BPD (P = 0.005), while the NR coregulator subset and complete NTC gene set showed association to BPD (P = 0.003) and SZ (P = 0.033) [Figure 2 and Supplementary Table 9]. While not taking into the account the significant genetic overlap between PDs[5], these associations remained significant for MDD and BPD even after adjusting for multiple testing by applying a conservative Bonferroni correction [Figure 2]. When we applied the same approach to summary statistics from non-PD GWASs where NR-mediated signaling has been reported to play a role[97-104], a very significant association was seen for the NR coregulator subset in height and BMI, as well as a moderate significant association of the NR coregulator subset to HF (Supplementary Figure 1; P = 0.002). For COVID-19 (positive vs. population), in which NR biology plays no obvious role, no association was observed [Supplementary Figure 1].
Figure 2. MAGMA gene set association analysis of the NTC gene set with separate analyses for the NR and NR coregulator subsets using summary statistics from large recent GWASs on the psychiatric disorders: attention deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD), major depressive disorder (MDD), bipolar disorder (BPD), schizophrenia (SZ), and cross disorder (CD). Black/red dotted lines mark nominal/Bonferroni-adjusted significance cut-off.
Whereas common variants of small effect contribute to all PDs[105], particularly early onset disorders, such as ASD, are enriched with RCVs[106]. To assess the genetic burden of NTC RCVs in PDs, we focused on large (> 3000 cases and controls) WES studies that have been conducted in SZ[77] and ASD[78]. In these studies, PD-associated RCVs were identified in SZ (a single gene) and ASD (102 genes). Strikingly, 19% of genes with ASD-associated RCVs are NTC-encoding genes [Supplementary Table 8], representing a significant overrepresentation [chi-squared test (one-tailed), P < 0.0001]. Furthermore, 32% of identified ASD-associated NTC RCV-harboring genes reside in PD GWS loci (e.g., RORB and FOXP1), thus supporting the pathoetiological relevance of these particular NTC genes within multi-gene GWS loci.
Patient epigenetic signature and brain transcriptomic profile support the implication of dysregulated nuclear receptor-mediated signaling in mental illness
Complementing genome-wide studies of DNA sequence variation, studies of variation to the epigenome have the potential to reveal biosignatures associated with disease-causing factors in mental illness[107]. Particularly, methylome-wide association studies (MWASs) have revealed hundreds of DNA methylation changes associated with PDs and psychiatry-related traits[80-84,108-114]. The epigenome is dynamic, changes in response to environmental[115] as well as endogenous factors (e.g., hormonal transitions[116,117] and aging[118]), and plays a crucial role in the orchestration of gene transcription in the developing human brain[86]. Clinical MWASs in brain tissues are rare and of small sample sizes. Hence, we assessed the burden of genes with changes in DNA methylation associated with PDs among the NTC gene set in the current largest patient blood MWASs. From neonatal samples, data were available for ADHD and ASD, where ~16% of findings with differential methylation were annotated to NTC genes [Supplementary Table 5 and Supplementary Table 9; chi-squared test (one-tailed), ASD: P = 0.001 and ADHD: P = 0.050]. From adults, samples have been collected and analyzed in ADHD, MDD, and SZ cases. Whereas none of the two differentially methylated genes identified in ADHD encode NTC genes, ~7% of differentially methylated genes in MDD belonged to the NTC [Supplementary Table 5 and Supplementary Table 8; chi-squared test (one-tailed), P = 0.024]. A similar overlap (~5% and 10%) was seen in two independent studies in SZ cases [Supplementary Tables 5 and 8; chi-squared test (one-sided), P = 0.037[79] and P < 0.0001[80]], whereas meta-analyses of SZ MWASs using a more stringent significance cut-off did not find NTC genes among 10 differentially methylated genes[81] [Supplementary Tables 5 and 8]. Notably, several differentially methylated NTC genes harbor ASD RCVs or reside in PD GWS loci (e.g., GATAD2A, RERE, CREBPB, and FOXP1), and several NTC genes were differentially methylated in more than one dataset/disorder (FOXP1, EP400, TRERF1 and SKI) [Figure 3 and Supplementary Tables 5 and 8]. Interestingly, data from a large MWAS of epigenetic plasticity during early fetal brain development reveal that > 40% of NTC genes undergo dynamic DNA methylation changes during early fetal brain development [Supplementary Table 5], thus supporting an important and meticulously orchestrated role for the NTC in transcriptional regulation in the developing human brain. NR-mediated signaling, however, remains important throughout life and altered cerebral expression of NR encoding genes have been reported in adult SZ cases[119]. To explore the transcriptomic signature of NTC genes in brain tissue from PD cases, we examined data from a comprehensive brain whole-transcriptome study conducted on postmortem dorsolateral prefrontal cortex (DLPFC) samples from 258 SZ patients and 271 healthy controls[92]. While only a minor fraction of NTC genes (PRKDC, PSMD1, AKAP13, IDE, SMAD3, HR, GADD45A, RBFOX2, and LCORL) were differentially expressed in SZ cases compared to healthy controls [Supplementary Figure 2], a quantitative analysis of promotor HREs in DEGs compared to genes displaying no regulation in cases revealed a nominally significant enrichment of RXRβ (P = 0.003), RORγ (P = 0.036), PR (P = 0.038), and HNF4α (P = 0.048) HRE sets in upregulated DEGs, and RORγ (P = 0.026), RXRα (P = 0.028), and RARγ (P = 0.049) HRE sets in downregulated DEGs [Supplementary Table 10].
Figure 3. MAGMA analyses of HRE gene set association using summary statistics from ADHD, ASD, MDD, BPD, SZ, and cross disorder (CDG2) GWAS as well as from a range of non-psychiatric GWASs [Alzheimer’s disease (AD), type 2 diabetes (T2D), body mass index (BMI), heart failure (HF), height, and COVID-19]. The red dotted line marks Bonferroni-adjusted significance cut-off. ADHD: Attention deficit/hyperactivity disorder; ASD: autism spectrum disorder; HRE: hormone response element; MDD: major depressive disorder; BPD: bipolar disorder; SZ: schizophrenia.
Psychiatric disorder risk enrichment among genes containing NR binding motifs
Both NTC members and NR ligands have been associated with PDs, but the contribution of their respective genomic actions in relation to PD risk is poorly understood. NRs bind to DNA as monomers, homodimers, and heterodimers, most commonly in a bimolecular complex with the retinoid X receptor (RXR)[120]. However, a recent study has demonstrated widespread binding of NRs to half-sites, and that half-site binding can drive transcription[121]. Hence, to assess the aggregated genetic burden in target genes of individual NRs, we used an in silico approach to test the gene set association of promotor HRE half-sites containing target genes of PD-associated NTCs using GWAS summary statistics. First, we assessed the association of HRE genes governed by NRs associated to PDs in GWASs or WESs. Whereas we did not see a significant association of RARE containing genes governed by SZ-associated RARγ, RORE gene set governed by CD-associated RORα was significantly associated with ASD (Figure 3, Table 1, and Supplementary Table 8; P = 0.022). Next, we profiled the risk landscape of HRE gene sets in general using summary statistics from both PDs and non-psychiatric traits. This revealed nominally significant associations of: ARE (P = 0.046) and FXRE (P = 0.049) gene sets with ADHD; PPARE (P = 0.045), FXRE (P = 0.023), RORE (P = 0.022), and NR1D1 targets (P = 0.046) with ASD; DAX1 target genes with SZ (P = 0.027); ERE (P = 0.042), GRE (P = 0.036), PGRE (P = 0.011), RARE (P = 0.047), and TRE (P = 0.031) with MDD; and ERE (P = 0.045), FXRE (P = 0.004), and RXRE (P = 0.042) with CD [Figure 3 and Supplementary Table 8]. In addition, a number of HRE gene sets showed association to non-psychiatric traits, including FXRE to BMI (P < 0.0001) and height (P < 0.0001). While the association between FXRE and BMI/height remained significant following a conservative Bonferroni correction for multiple testing, it is important to realize that NRs regulate distinct yet highly overlapping gene programs[121]. To assess the overlap of HRE gene sets, we assessed and plotted their pairwise similarities [Supplementary Figure 3 and Supplementary Table 11]. Not surprisingly, > 95% of HRE gene sets displayed a significant overlap of genes, with particularly closely related superfamily members displaying the highest degree of overlap in their target gene sets (e.g., GR and AR, ERα and ERβ, HNF4γ and HNF4α, and PXR and CAR), thus arguably reducing the number of effective independent tests performed.
Summary of the genetic/epigenetic associations of NTC genes to psychiatric disorders
| Family | Gene name | Gene symbol | Gene synonym | GWAS | WES | MWAS | GWAS catalogue | Interacting NR coregulators | HRE gene set association | |||
| GWAS | WES | MWAS | GWAS catalogue | |||||||||
| 0B | Short heterodimeric partner | NR0B2 | SHP | BBX (SZ), BRMS1 (BPD) | SIN3A (ASD) | |||||||
| 0B | Dosage-sensitive sex reversal-adrenal hypoplasia congenital critical region on the X chromosome, Gene 1 | NR0B1 | DAX1 | SZ | ||||||||
| 1A | Thyroid hormone receptor-α | NR1A1 | THRA | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa), NCOR2 (cocaine dependence), TBL1Y (ASD) | MDD | ||||
| 1A | Thyroid hormone receptor-β | NR1A2 | THRB | SZ, MDD in trauma-unexposed individuals, general cognitive ability, intelligence | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa), NCOR2 (cocaine dependence), TBL1Y (ASD) | ||||
| 1B | Retinoic acid receptor-α | NR1B1 | RARA | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa), NCOR2 (cocaine dependence), TBL1Y (ASD) | |||||
| 1B | Retinoic acid receptor-β | NR1B2 | RARB | Oppositional defiant disorder dimensions in ADHD | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | NCOR2 (cocaine dependence), TBL1Y (ASD) | MDD | |||
| 1B | Retinoic acid receptor-γ | NR1B3 | RARG | SZ | BPD or attention deficit hyperactivity disorder, personality traits in BPD | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa), NCOR2 (cocaine dependence), TBL1Y (ASD) | |||
| 1C | Peroxisome proliferator-activated receptor-α | NR1C1 | PPARA | SZ | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | NCOR2 (cocaine dependence), TBL1Y (ASD) | ||||
| 1C | Peroxisome proliferator-activated receptor-δ | NR1C2 | PPARD | MDD | Response to antipsychotic treatment | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | ||||
| 1C | Peroxisome proliferator-activated receptor-γ | NR1C3 | PPARG | EP300 (CD/SZ/MDD), NRIP1 (SZ) | CREBBP (ASD), NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa), NCOR2 (cocaine dependence), TBL1Y (ASD) | ASD | ||||
| 1D | Reverse-Erb-α | NR1D1 | ASD | |||||||||
| 1F | Retinoic acid receptor-related orphan receptor-α | NR1F1 | RORA | CD | General cognitive ability, SZ, educational attainment (MTAG), educational attainment (years of education), depression (quantitative trait), response to citalopram treatment | EP300 (CD/SZ/MDD) | ASD | |||||
| 1F | Retinoic acid receptor-related orphan receptor-γ | NR1F3 | RORC | Insomnia | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | ||||||
| 1F | Retinoic acid receptor-related orphan receptor-β | NR1F2 | RORB | SZ | ASD | Depressive symptoms (SSRI exposure interaction) | ||||||
| 1H | Liver X receptor-β | NR1H2 | LXRB | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa) | |||||
| 1H | Liver X receptor-α | NR1H1 | LXRA | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | |||||||
| 1H | Farnesoid X receptor-α | NR1H4 | FXRA | EP300 (CD/SZ/MDD), PRMT1 (SZ), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa) | CD, ASD, ADHD | ||||
| 1I | Vitamin D receptor | VDR | EP300 (CD/SZ/MDD), NRIP1 (SZ), NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa) | |||||||
| 1I | Pregnane X receptor | NR1I2 | PXR | NRIP1 (SZ) | NCOA1 (ASD) | MGMT (anorexia nervosa) | ||||||
| 1I | Constitutive androstane receptor | NR1I3 | CAR | NCOA1 (ASD) | ||||||||
| 2A | Hepatocyte nuclear factor-4-α | HNF4A | ||||||||||
| 2B | Retinoid X receptor-α | RXRA | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | MGMT (anorexia nervosa) | CD | |||||
| 2B | Retinoid X receptor-β | RXRB | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | ||||||||
| 2B | Retinoid X receptor-γ | RXRG | NCOA1 (ASD) | |||||||||
| 2C | Testicular orphan nuclear receptor 4 | NR2C2 | TR4 | NR2C2AP (BPD) | ||||||||
| 2F | Chicken ovalbumin upstream promoter-transcription factor-α | NR2F1 | COUP-TF1 | BCL11B (CD/SZ) | NCOA1 (ASD) | |||||||
| 2F | Chicken ovalbumin upstream promoter-transcript | NR2F2 | COUP-TF2 | BCL11B (CD/SZ) | ||||||||
| 2F | V-Erb-A avian erythroblastic leukemia viral oncogene homolog-like 2 | NR2F6 | NCOA1 (ASD) | |||||||||
| 3A | Estrogen receptor-β | ESR2 | CD/MDD | Educational attainment (years of education), depression | EP300 (CD/SZ/MDD) | NCOA1 (ASD) | NCOR2 (SZ) | CD | ||||
| 3A | Estrogen receptor-α | ESR1 | Educational attainment (years of education), educational attainment (MTAG), alcohol dependence, developmental language disorder, anxiety | EP300 (CD/SZ/MDD), SRA1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | MDD | |||||
| 3B | Estrogen-related receptor-β | ESRRB | ASD spectrum disorder, attention deficit hyperactivity disorder symptoms (maternal expressed emotions interaction) | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | MGMT (anorexia nervosa) | ||||||
| 3B | Estrogen-related receptor-γ | ESRRG | Major depression and alcohol dependence, alcohol consumption, cognitive aspects of educational attainment, cognitive performance, cognitive performance (MTAG), general cognitive ability, intelligence, intelligence (MTAG), major depressive disorder, adventurousness | EP300 (CD/SZ/MDD), TLE1 (SZ), NRIP1 (SZ) | NCOA1 (ASD) | |||||||
| 3B | Estrogen-related receptor-α | ESRRA | NRIP1 (SZ) | NCOA1 (ASD) | MGMT (anorexia nervosa), NCOR2 (cocaine dependence), TBL1Y (ASD) | |||||||
| 3C | Androgen receptor | AR | EP300 (CD/SZ/MDD), KAT5 (SZ), SMARCD1 (SZ), BCL7A (SZ), NRIP1 (SZ) | NCOA1 (ASD), ARID1B (ASD), SMARCC2 (ASD) | NCOR2 (SZ) | |||||||
| 3C | Mineralocorticoid receptor | NR3C2 | MR | ASD | SZ, well-being spectrum (multivariate analysis), benign childhood epilepsy with centro-temporal spikes | EP300 (CD/SZ/MDD), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (cocaine dependence), TBL1Y (ASD) | ||||
| 3C | Glucocorticoid receptor | NR3C1 | GR | Night sleep phenotypes | EP300 (CD/SZ/MDD), SMARCD1 (SZ), BCL7A (SZ), NRIP1 (SZ) | NCOA1 (ASD), ARID1B (ASD), SMARCC2 (ASD) | NCOR2 (SZ) | MDD | ||||
| 3C | Progesterone receptor | NR3C3 | PGR | EP300 (CD/SZ/MDD), SRA1 (SZ), NRIP1 (SZ) | NCOA1 (ASD) | NCOR2 (SZ) | NCOR2 (cocaine dependence), TBL1Y (ASD), TBL1Y (ASD) | MDD | ||||
| 4A | Nerve growth factor 1B | NR4A1 | NGFI-B | EP300 (CD/SZ/MDD) | NCOA1 (ASD) | |||||||
| 4A | Neuron-derived orphan receptor-1 | NR4A3 | NOR-1 | EP300 (CD/SZ/MDD) | ||||||||
| 4A | Nurr-related factor 1 | NR4A2 | NURR1 | MDD | ||||||||
| 5A | Steroidogenic factor-1 | NR5A1 | SF1 | NCOA1 (ASD) | ||||||||
| 6A | Liver receptor homolog-1 | NR5A2 | LRH1 | General cognitive ability, nicotine dependence | EP300 (CD/SZ/MDD) | NCOA1 (ASD) | ||||||
Brain-transcriptomic profile of the nuclear receptor transcriptome complex hints at a neurodevelopmental impact of psychiatry-associated nuclear receptor networks
The transcriptional activity of NRs critically depends on their interactions with NR coregulators. The biophysical interactions have been established in vitro between a range of NRs and NR coregulators[49] (see Supplementary Table 6 for a list of well-documented interactions), but the biological relevance of these interactions in the brain depends on their co-expression in the same structures and individual brain cells. Hence, we assessed single cell expression characteristics of NTC genes and identified gene sets that are specific to individual brain cell types[95]. Overall, 23% of NRs and 13% of NR coregulators are exclusively expressed in specific brain cell types
Next, we clustered NTC genes based on co-expression characteristics in the developing human brain [Figure 4]. This revealed eight distinct larger co-expression clusters each characterized by peak expression in specific developmental stages or tissues. While the majority of NR encoding genes peak postnatally (Figure 4 and Supplementary Table 8; Clusters 1-6), a subset (NURR1, NOR-1, NR5A2, TR4, COUP-TF1, COUP-TF2, RORB, THRA, RARA, and ESR2) peak at the earliest stage of development (Figure 4 and Supplementary Table 8; Cluster 7). Within this group, COUP-TF1 and -2 are particularly abundantly expressed in the amygdala, while NOR-1 expression peaks in the hippocampus [Figure 4]. Interestingly, the cluster of NTC genes peaking prenatally hosts the highest density of genes in PD GWS loci and ~80% of RCV harboring NTC genes associated with the early onset PD, ASD. It is also interesting that a cluster of 23 NTC genes is predominantly expressed in striatal tissue, with a subset displaying very high expression in prenatal striatal tissue. This striatal-dominant cluster includes the NR encoding genes RARB, RXRG, and SF1, as well as FOXP1 identified in both SZ MWAS and ASD WES (Figure 4 and Supplementary Table 8; Cluster 4). A third cluster with peak expression in the cerebellum houses nine NR encoding genes (ESRRA, NR2F6, RARG, RORC, RXRB, SHP, ESRRG, RORA, and ESRRB), of which the CD GWS RORA along with ESRRA and ESRRG display particularly high expression in the prenatal cerebellum (Figure 4 and Supplementary Table 8; Cluster 1). A summary of brain cell-specific, co-expressed NTC genes is presented in Supplementary Figure 5.
Figure 4. Expression of NR and NR coregulator encoding genes across 16 brain structures and four developmental stages. Row annotations include: NTC subtype (NR/NR coregulator) and genes in GWAS/WES/MWAS loci. Genes fall in eight major clusters defined by most abundant expression in: (1) cerebellum; (2) adults; (3) puberty and adulthood; (4) striatum; (5) prenatally and in childhood; (6) childhood; (7) prenatally; and (8) prenatally and puberty. A1C: Primary auditory cortex; AMY: amygdala; CBC: cerebellar cortex; DFC: dorsolateral prefrontal cortex; HIP: hippocampus; ITC: inferolateral temporal cortex; M1C: primary motor cortex; MD: mediodorsal nucleus of thalamus; MFC: anterior cingulate cortex; OFC: orbital frontal cortex; S1C: primary somatosensory cortex; STC: posterior superior temporal cortex; STR: striatum; V1C: primary visual cortex; VFC: ventrolateral prefrontal cortex.
Sex-biased expression of the nuclear receptor transcriptome complex in the developing and mature human brain
Sex differences are common in PDs where symptom profiles and severity differ between men and women[20-25], and, e.g., women are more susceptible to affective disorders than men[24,25]. Brain development follows sex differential trajectories[122] with concordant regional sex-biased expression of comprehensive gene sets. Sex hormones act throughout the brain of both men and women, but subtle differences exist in their genomic and non-genomic actions[123]. Sex-biased expression of the ASD candidate and CD GWS annotated gene, RORA, has been suggested as a contributor to the sex-bias in ASD[124]. We speculated that sex-biased expression of NTC genes in general contribute to the sex-biases in mental illness. Hence, we assessed the overlap between the NTC gene sets and reported sex-DEGs across brain regions at four developmental stages (prenatal, early childhood, puberty, and adulthood)[96].
Whereas we did not find significant enrichment of NTC genes overall, we found that sex-biased genes are significantly enriched with NR encoding genes at the prenatal stage, with particular enrichment among sex-biased genes in the medial frontal cortex (P = 0.006), orbitofrontal cortex (P = 0.004), and striatum (P = 0.010) [Figure 5 and Supplementary Table 12]. In frontal cortical tissues, these include RORB, NR4A2, NR4A3, and NR3C2, the expression of which are all higher in women than in men. In striatal tissue, NURR1, NR1D2, NR2F6, and THRB are all sex-differentially expressed with expression being higher in men - except for NR2F6 that is female-biased. Several NR coregulators are similarly sex-differentially expressed in these structures in the prenatal stage. At later developmental stages, expression of sex-biased NTC genes is consistently higher in women compared to men [Figure 5]. Interestingly, NTC genes in SZ GWS loci are significantly overrepresented among NTC genes that display male-biased expression in the prenatal striatum (Fisher’s exact test; P = 0.0193) and NTC genes with ASD-associated RCVs among female-biased genes in the prenatal orbito- and medial frontal cortex (Fisher’s exact test; P = 0.003). The density of PD-associated NTC genes were furthermore high in the cluster of genes with female-biased cortical expression in puberty [Figure 5]. Among genes reported to be sex-differentially methylated in the earliest stages of fetal brain development, only a minor fraction encodes NTC genes [Supplementary Table 5], but particularly SZ MWAS risk genes clustered among genes with female-biased thalamic dominant expression [Figure 5].
Figure 5. Sex-biased expression of NTC genes. Mapped are NTC genes that are sex-differentially expressed in the developing human brain as reported by Shi et al.[96]. Red indicates higher expression in females, while blue indicates higher expression in males. Common and rare variants in NTC genes associated with particularly SZ and ASD are enriched in clusters with female-biased expression in the anterior cingulate cortex (MFC) and orbital frontal cortex (OFC) and male-biased expression in striatum (STR) during prenatal development. SZ MWAS risk genes cluster with genes expressed with female-biased MD (mediodorsal nucleus of thalamus) in childhood. NTC: NR transcriptomic complex; SZ: schizophrenia; ASD: autism spectrum disorder.
DISCUSSION
Human brain development is a protracted process that begins in the early prenatal stage and extends through late adolescence and even adulthood[125]. The process is genetically organized, but it is shaped and adapted in the context of environmental input. Neither genes nor environmental clues are determinative in terms of outcome, but disruption to either may affect the maturing brain and mind. The CNS and the endocrine system work in synergy to sense and act upon endogenous and environmental cues. Whereas the CNS response is rapid and mostly transient, the endocrine response maintains homeostasis and long-term control through various molecular mechanisms that include the genomic actions of ligand-activated NRs. In line with the scientific consensus that the origin of psychopathology is neurodevelopmental, the brain is most vulnerable to the effect of steroid imbalances and disrupted NR-mediated signaling at the earliest stages of development[126-129]. Balanced NR-mediated signaling, however, remains important throughout life, and steroid levels exhibit a maximum in young men and women (~20 years)[130] but vary greatly in abundance during periods of hormonal transition (childhood, puberty, post-partum, and menopause), thus overlapping with the vulnerability periods and age of onset of many PDs. Altered steroidogenic activity and imbalances in total circulating cholesterol and other lipid metabolites have been reported in a range of PDs[131,132]. In addition to endogenous steroids and derivatives of retinoids, fatty acids, cholesterol, lipophilic hormones, and vitamins, NRs further act as sensors for a range of xenobiotics, antibiotics, and synthetic compounds[16] - with implications for the therapeutic effect of CNS drugs and CNS side effects of non-CNS-targeting drugs[133]. NR-mediated signaling thus constitutes a delicate molecular mechanism that is both vulnerable to biological dysregulation and interesting as a pharmacological target in the context of mental health.
Genomic vulnerability to dysregulated nuclear receptor-mediated signaling in mental illness
We found that the genetic NTC risk burden is high across psychiatric diagnostic entities. Particularly, we found that on average ~15% of SZ, MDD, and BPD GWS loci harbor NTC genes, and the NTC gene set overall displays significant association to these disorders. In addition, nearly 20% of ASD-associated RCV-harboring genes are members of the NTC. Although genetic studies have highlighted the implication of individual NR and NR coregulator-encoding genes in mental illness, this is the first study to demonstrate a consistently elevated genetic burden in the NTC in PDs. The biological relevance of this overrepresentation of NTC genes among PD risk genes is further substantiated by the high number of NTC genes that reside in multi-PD and CD GWS loci and the enrichment of NTC genes among differentially methylated genes in PDs. We note that particularly the NR subset of the NTC is associated with affective disorders, whereas the risk burden in NR coregulators is dominant in SZ and ASD. Interestingly, NR-encoding genes generally peak in their expression postnatally, whereas NR coregulators - particularly those associated with ASD and SZ - peak in their expression at the earliest stages of brain development. This may be related to the differences in onset between affective and non-affective PDs. In this regard, it is interesting to note that ASD risk NTC genes cluster among genes that display female-biased expression in the prenatal cortex and male-bias in the prenatal striatum, while non-SZ GWS NTC genes, on the contrary, overlap with genes that are male-biased in the prenatal cortex and female-biased in the postnatal thalamus and cortex. It is thus conceivable that differences in baseline NTC gene expression in males and females impact on their vulnerability to genetic alterations in these gene sets and, consequently, on their sex-biased PD risk profiles.
PD-associated NRs are not restricted to the endocrine receptor subclass of the NR family, but they include lipid sensors and adopted and true orphan receptors, thus potentially broadly bridging the gap between genetic and epidemiological risk. In line with this notion, many PD-associated NR coregulators are ubiquitously expressed in the brain and share a broad range of interactions with PD-associated NRs
Figure 6. NR-signaling pathways with high genetic risk burden. Illustrated are individual NRs and their experimentally validated associated NTC, their HRE target genes, resulting transcripts, selected ligands, and linked epidemiological risks, as well as selected psychiatry-relevant drugs. The illustration is divided into experimental levels (epidemiological, molecular, genetic, and pharmacological) as well as the developmental stage/brain structure in which the individual NR-encoding genes peak in expression. Entities associated with SZ are highlighted in blue, MDD in green, ASD in grey, CD in orange, and multiple PDs in black. NTC: NR transcriptomic complex; HRE: hormone response element; NR: nuclear receptor; SZ: schizophrenia; MDD: major depressive disorder; ASD: autism spectrum disorder; CD: cross-disorder; PDs: psychiatric disorders.
The functional output of signaling through NRs is a change in transcription of gene sets containing promotor HRE sequences. Whereas we did not find a strong transcriptomic NTC signature in postmortem brain samples from adult SZ cases, the enrichment of particular HRE sequences in the promotors of DEGs is in agreement with altered cerebral NR-mediated signaling in SZ. However, it is important to note that many commonly administered drugs in psychiatry and comorbid disorders will affect CNS NR-mediated signaling. Hence, it is not possible to ascribe the observed enrichment to a biological disorder or treatment.
At the genomic level, we found that some HRE-containing gene sets are associated with individual PDs, whereas others display association to PDs in general. This includes the HRE target genes of gonadosteroid receptors (PGR and ERα) and the retinoic acid receptor (RARβ), which are exclusively associated with MDD, and RORα HRE targets which have no association to PDs besides ASD. On the other hand, the retinoic acid receptor X α (RXRα) target genes appear to be more generally associated with mental illness, in line with the role of RXR heterodimeric complexes[120].
Supporting the biological relevance of the observed associations, subsets of HRE gene sets displayed association to diseases in which NRs are reportedly involved. These include the association of target genes of the NR1I subfamily of NRs (PXR and CAR that are generally implicated with regulation of energy metabolism and insulin sensitivity[135,136]) and the phenotypically interlinked diseases/traits: T2D, HF, and BMI. PPARs have been associated with T2D[137] and AD[100]. Interestingly, the VDRE gene set was significantly associated with AD and T2D in line with the reported associations between low serum 25-hydroxyvitamin D levels and AD and T2D[138], but not with, e.g., ASD and SZ that have been associated with early life vitamin D deficiency[17,19]. RXRE was nominally significantly associated with AD, where RXR agonist administration leads to significant decrease in brain amyloid burden[139]. On the contrary, no association was observed between HRE gene sets and COVID-19 (positive vs. population), where NR biology plays no obvious biological role.
Genetic, epidemiological, empirical, and pharmacological evidence highlight distinct psychiatry-relevant nuclear receptor-mediated signaling pathways
Cell type- and tissue-specific co-expression is required for biophysical assemblage and psychiatry-relevant genomic signaling by distinct NTCs. We clustered NTC genes based on their co-expression characteristics in the developing human brain and identified networks of putative cell-specific NTCs with meticulously documented interactions. This revealed NTCs of known biological relevance, as well as novel NTCs with putative pharmacological potential in psychiatry. Here, we highlight selected NTCs whose implication in PDs are supported by multilevel genomic and known epidemiological, empirical, and pharmacological evidence.
Estrogen, androgen, and progesterone receptors
Gonadosteroid-binding receptors are among the NRs with implications in PDs supported by strong and multilevel evidence. Women who are in their peak estrogen-producing years or transitioning to menopause are at an elevated risk of developing affective disorders, as are women who are experiencing hormonal fluctuations, e.g., during menstrual periods and postpartum[140]. Sex-biases characterize PDs in general, and altered levels of progesterone and androgens have been reported in SZ and estrogen levels in numerous PDs[141]. In addition, hormone replacement therapy has successfully been used in the treatment of PDs, including MDD, BPD, ASD, ADHD, and SZ[35], with positive outcomes of testosterone replacement therapy in MDD[142]. Estrogen replacement therapy has been successful in postpartum depression[142] and has demonstrated antimanic effects in women with BPD (tamoxifen and raloxifene)[143] and improvement of positive and negative symptoms in SZ patients[35,144]. At the genetic level, the estrogen receptor-encoding gene (ESR2) resides in a GWS locus associated with both CD and MDD, and older association studies have repeatedly implicated ESRs with a range of PDs and psychiatry-related traits. Convincingly, the ESRE target gene set of ERβ is similarly associated with CD, thus strongly supporting a pathobiological relevance of imbalanced genomic ERβ signaling in mental illness at a broader level. Interestingly, ESR2 locates to a different co-expression cluster than the genes encoding the other gonadosteroid-sensing receptors (ERα, AR, and PGR). Particularly, the ESR2 gene cluster peaks prenatally, while the others peak during puberty and adulthood. However, ERα and PGR both display a link to MDD, as their target ESRE and PGRE gene sets are both associated with MDD [Figure 6]. DEGs identified in SZ postmortem brains are further enriched with PGRE in their promotor sequences. All gonadosteroid receptors share a range of NR coregulators, but both ESR1 and PGR show cell-specific expression (neurons and endothelia, respectively) and have potential receptor-specific NR coregulators from within their co-expression clusters. For ERα, this includes SZ GWS PRMT8, although their biophysical interaction remains to be systematically examined.
Corticosteroid receptors
Exposure to traumatic, maternal, and early life stress is a major risk factor in many PDs, including SZ, BPD, MDD, and anxiety disorders[26,145,146]. Among the NTC genes that harbor ASD-associated RCVs is NR3C2 encoding the mineralocorticoid receptor (MR). MR is a high-affinity corticosteroid receptor that acts in synergy with the glucocorticoid receptor (GR) to mediate the molecular stress response. Both GR and MR belong to a gene co-expression cluster with peak expression in puberty and adulthood; however, whereas GR is widely expressed in the brain and peak in cerebellar tissue, MR expression peaks in limbic tissues, in accordance with previously published reports[147]. MR plays a well-documented and sex-biased role in stress resilience and depression[148], where a functional MR haplotype protects against depression following early life trauma[149]. Unlike MRE target genes, the GRE gene set showed a significant association to MDD [Figure 6]. This is in line with a recent study that demonstrated that genetic differences in the immediate transcriptome response to stress predict the risk of several PDs[18].
Retinoid binding nuclear receptor
Retinoids play a crucial role in developmental pathways, but they are also essential to a number of postnatal processes, including synaptic plasticity[28]. Retinoid signaling is mediated through binding to RARs and PPARs in heterodimeric partnership with RXR. Low maternal retinol is a risk factor in SZ in adult offspring 29, and membrane levels of several PUFAs, which signal through the same receptors[150,151], have been associated with psychotic, depressive, and manic symptoms in individuals at ultrahigh risk for psychosis[39]. Accumulating evidence has implicated retinoid signaling in the pathoetiology of particularly SZ (recently reviewed by Reay et al.[28]), and PPAR/RXR and RAR/RXR complexes have been proposed as therapeutic strategies in CNS disorders[152]. Among the RXRs, none have been found in PD GWS loci; however, we found that the RXRE target gene set of RXRα is significantly associated with CD. Whereas RXR-encoding genes are not restricted to specific cell types, their heterodimeric partners, PPARs, are. PPARγ is specific to microglia, PPARα to astrocytes, and PPARδ to endothelia among brain cells - and both PPARA and PPARG are co-expressed with PD-associated coregulators in these specific cells. While none of the three receptors have been associated with PDs in GWASs, PPARA and PPARD are differentially methylated in blood from, respectively, SZ and MDD patients. In addition, we found that the PPARE target gene set of PPARγ is significantly associated with ASD. It is further noteworthy that both PPARα and -γ can bind and respond to cannabinoids[153], thus providing a potential genetic link to the risks and phenotypes associated with cannabis use in PDs[154].
Among the RAR encoding genes, only RARG resides in a PD GWS locus (SZ), whereas we found that the RARE target gene set of RARβ is associated with MDD. RARB and RARG have different expression profiles, and, whereas RARG clusters with genes with peak expression in the cerebellum, RARB expression peaks in striatal tissue. RARβ and RARγ share a number of NR coregulator interaction partners genetically associated with PDs [Figure 6].
RORs
Patients with pathogenic variations in retinoic acid receptor-related orphan receptors (RORs) present with ASD as well as seizures. Both RORs (RORα and RORβ) are located in PD GWS loci. The RORB gene is associated with SZ and further harbors ASD-associated RCVs. RORB is specifically expressed in astrocytes and resides in a gene co-expression cluster that peaks during prenatal brain development. Besides RORB, none of the NTC genes with prenatal peaks are astrocyte specific, but single cell genomics in ASD cortical tissue have associated altered glial RORB expression with ASD[155]. Further supporting the involvement of ROR-mediated signaling in ASD, RORα resides in a CD GWS locus and its RORE-containing target genes are significantly associated with ASD [Figure 6]. This is in agreement with reported ASD risk genes under RORα transcriptional regulation[156]. Similarly, an association has been found between ASD and the significantly overlapping HRE-containing target genes of NRID1 (Rev-ErbA-Alpha) that reportedly acts as a repressor of RORE gene sets[157]. Reduced RORA transcript and/or protein levels has been reported in both blood and postmortem brain tissue from ASD cases[158]. RORs are involved in a number of psychiatry-relevant pathways including neurogenesis, stress response, and modulation of circadian rhythms[159]. RORα binds with high affinity to the brain-specific cholesterol-metabolite, 24S-hydroxysterol (cerebrosterol), which has been found differentially abundant in plasma and suggested as a biomarker in ASD[160].
Orphan receptors
Located in an MDD GWS locus, NURR1 is specifically expressed in microglia and co-expressed with the SZ GWS NR coregulators, CNOT1 and GMEB1. However, the biophysical interaction of these coregulators with NURR1 has not been systematically examined. Although classified as an orphan receptor, NURR1 activity can be modulated by several small molecules [including docosahexaenoic acid (DHA) and other unsaturated fatty acids][161], as well as non-steroidal anti-inflammatory drugs[162]. NURR1 has been characterized as a neuroprotective and anti-inflammatory transcription factor[163] and suggested as a therapeutic target in Parkinson’s disease[164]. The monomer NBRE targets of NURR1 are not significantly associated with any PD, but, as NURR1 can bind DNA as a heterodimer with RXRs, it has the potential to modulate CD-associated RXRE target genes.
Little is reported about a role for the DAX1 receptor in mental illness. It is an orphan receptor and has been reported to act as a repressor of other NRs through heterodimeric interactions with, e.g., MR and GR[165,166]. However, in the brain, DAX1 is specifically expressed in oligodendrocytes. We found that the HRE half-site targets of DAX1 display significant association with SZ and interact with several PD-associated NR coregulators [Figure 6].
Therapeutic potential of targeting nuclear receptor biology in psychiatry
The activity of NRs can be pharmacologically modulated by specific ligands, thereby allowing for agonism, partial agonism, and antagonism. This has made them primary therapeutic targets for decades[167], and approximately 16% of FDA approved drugs target NRs[168]. A wide spectrum of somatic disorders has successfully been targeted by drugs directed at NRs. PPARγ-targeting thiazolidinediones are used in the treatment of diabetes, cardiovascular disease, and cancer[169]; selective ER modulators in ER-positive and metastatic breast cancer[170]; and RXR/RAR-targeting isotretinoin against acne. Furthermore, the well-known drug bexarotene, a selective RXR agonist, has been effectively used in the treatment of cutaneous T-cell lymphoma. A range of NR-targeting drugs have also proven efficient in non-psychiatric traits of the CNS, although most have yet to demonstrate clinical efficacy and sustainability in phase III trials. Whereas NR modulators are increasingly recognized as potentially powerful therapeutics for neurodegenerative CNS diseases[104,171-174], a similar shift in focus remains to be seen for drug discovery programs in PDs. NRs have been suggested as therapeutic targets in PDs[175], and pharmacological targeting of NR-mediated signaling has demonstrated clinical efficacy in the treatment of PDs[176], as assessed following administration of thyroid hormones (liothyronine), progesterone receptor antagonist (mifepristone), and bexarotene in affective disorders and SZ, respectively[177,178].
Despite their positive effects, but likely owing to their wide applicability, many drugs targeting NRs are associated with serious adverse effects[169,170], affecting also the CNS - for instance, suicidal behavior following administration of the widely prescribed acne-drug Accutane (isotretinoin)[179]. Other NR-targeting therapeutic strategies completely fail to demonstrate clinical efficacy, in which cases poor penetration of the blood-brain barrier seems to be the main impediment. Interestingly, a recently developed fatty acid amide hydrolase (FAAH)-targeting prodrug strategy appears to successfully facilitate blood-brain barrier diffusion through masking of small molecule carboxylate-containing NR modulators of therapeutic relevance to CNS disorders including ligands for TR, RXR, PPAR, LXR, ER, and RAR[42].
NRs are extensively expressed throughout the brain, in many tissues and cell types, making them particularly difficult to target without side effects. In the wake of this realization, accumulating interest has risen for the targeting of NR coregulators, which tend to be restricted to certain regions and cell types of the brain. Although commonly viewed as “undruggable” targets due to their large and flexible structures, potent small-molecule drugs have been developed to overcome this obstacle[180]. Other drugs target NR coregulators in an indirect manner through direct interaction with their NR, modulating the interaction between coregulator and NR, and thus the regulation of target genes[181,182]. We showed that both the NR and NR coregulator components of the NTC are overrepresented among PD risk genes, supporting the biological relevance of targeting this group of endogenous coregulators in psychiatry.
Here, we provide a resource for targeting psychiatry-relevant NTC networks with narrow cell specificity and defined sets of co-expressed interaction partners, which may significantly constrain the burden of off-target effects, favoring drug precision and safety in NR-based CNS therapeutics.
Perspectives and future research directions
There is an urgent need to identify molecular mechanisms implicated in PDs in order to progress the development of improved diagnostic tools and personalized medicine in psychiatry. Through mining of large-scale genomics data, we uncovered an unacknowledged genetic burden in NTC genes and their downstream genomic targets, supporting dysregulated NR-mediated signaling as a common and core molecular pathway in PDs. It is thus conceivable that NRs bridge the gap between genetic and epidemiological risk in mental illness, and that the genetic burden on associated molecular pathways may direct the individual’s vulnerability to adverse exposures and predict their clinical risk profile. This holds potential for both drug discovery and options in terms of molecular diagnostics and patient stratification. NR-mediated signaling has been suggested as a therapeutic target in PDs[175], but, due to the complexity of the NR interaction network, it is challenging to target specific functions of the network while avoiding serious adverse effects. The mechanisms by which individual cells modulate tissue-specific psychiatry-relevant NR ligand responsiveness is thus a fundamental issue in targeting NR-mediated signaling in the brain. Here, we categorized the genetic and epigenetic NTC risk burden in clusters of cell-specific and co-expressed genes that may provide a useful framework for future CNS NR therapeutic strategies in psychiatry.
DECLARATIONS
AcknowledgmentsWe thank the International Genomics of Alzheimer's Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i-Select chips were funded by the French National Foundation on Alzheimer's disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer's Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01-AG-12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer's Association grant ADGC-10-196728.
Authors’ contributionsWrote the manuscript: Donskov JG, Qvist P
Performed the experiments: Donskov JG, Starnawska A, Pallesen J, Qvist P
Performed the statistical analyses: Donskov JG, Starnawska A, Pallesen J, Qvist P
Conceived the basic idea and initiated the study: Qvist P
Designed and directed the study: Grove J, Børglum AD, Qvist P
All authors have read and approved the final manuscript.
Availability of data and materialsNot applicable.
Financial support and sponsorshipThis study was funded by the Lundbeck Foundation, Denmark (https://lundbeckfonden.com/; grant number R155-2014-1724) (Børglum AD), Aarhus University, Department of Biomedicine (Donskov JG), and the Augustinus Foundation (https://augustinusfonden.dk/) (Qvist P).
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
Supplementary MaterialsREFERENCES
1. Lee SH, Ripke S, Neale BM, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium; International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013;45:984-94.
2. Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013;381:1371-9.
4. Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 2019;179:1469-82.e11.
5. Smeland OB, Bahrami S, Frei O, et al. Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry 2020;25:844-53.
6. Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry 2014;55:1068-87.
7. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421-7.
8. Wray NR, Ripke S, Mattheisen M, et al. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 2018;50:668-81.
9. Grove J, Ripke S, Als TD, et al. Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; BUPGEN; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; 23andMe Research Team. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019;51:431-44.
10. Stahl EA, Breen G, Forstner AJ, et al. Bipolar Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 2019;51:793-803.
11. Demontis D, Walters RK, Martin J, et al. ADHD Working Group of the Psychiatric Genomics Consortium (PGC); Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium; 23andMe Research Team. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 2019;51:63-75.
12. Uher R, Zwicker A. Etiology in psychiatry: embracing the reality of poly-gene-environmental causation of mental illness. World Psychiatry 2017;16:121-9.
13. Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet 1985;19:209-52.
14. Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell 1995;83:835-9.
17. Eyles DW, Trzaskowski M, Vinkhuyzen AAE, et al. The association between neonatal vitamin D status and risk of schizophrenia. Sci Rep 2018;8:17692.
18. Arloth J, Bogdan R, Weber P, et al. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (PGC); Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium PGC. Genetic differences in the immediate transcriptome response to stress predict risk-related brain function and psychiatric disorders. Neuron 2015;86:1189-202.
19. Lee BK, Eyles DW, Magnusson C, et al. Developmental vitamin D and autism spectrum disorders: findings from the Stockholm Youth Cohort. Mol Psychiatry 2021;26:1578-88.
20. Zagni E, Simoni L, Colombo D. Sex and gender differences in central nervous system-related disorders. Neurosci J 2016;2016:2827090.
22. McLean CP, Asnaani A, Litz BT, Hofmann SG. Gender differences in anxiety disorders: prevalence, course of illness, comorbidity and burden of illness. J Psychiatr Res 2011;45:1027-35.
23. Riecher-rössler A. Oestrogens, prolactin, hypothalamic-pituitary-gonadal axis, and schizophrenic psychoses. Lancet Psychiatry 2017;4:63-72.
25. Kuehner C. Why is depression more common among women than among men? Lancet Psychiatry 2017;4:146-58.
26. Heim C, Newport DJ, Mletzko T, Miller AH, Nemeroff CB. The link between childhood trauma and depression: insights from HPA axis studies in humans. Psychoneuroendocrinology 2008;33:693-710.
27. Shi L, Lu ZA, Que JY, et al. Prevalence of and risk factors associated with mental health symptoms among the general population in China during the Coronavirus Disease 2019 Pandemic. JAMA Netw Open 2020;3:e2014053.
28. Reay WR, Cairns MJ. The role of the retinoids in schizophrenia: genomic and clinical perspectives. Mol Psychiatry 2020;25:706-18.
29. Bao Y, Ibram G, Blaner WS, et al. Low maternal retinol as a risk factor for schizophrenia in adult offspring. Schizophr Res 2012;137:159-65.
30. Lane MA, Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol 2005;75:275-93.
32. Stanikova D, Luck T, Bae YJ, et al. Increased estrogen level can be associated with depression in males. Psychoneuroendocrinology 2018;87:196-203.
33. Huber T, Rollnik J, Wilhelms J, von zur Mühlen A, Emrich H, Schneider U. Estradiol levels in psychotic disorders. Psychoneuroendocrinology 2001;26:27-35.
34. Brzezinski-Sinai NA, Brzezinski A. Schizophrenia and sex hormones: what is the link? Front Psychiatry 2020;11:693.
35. Hwang WJ, Lee TY, Kim NS, Kwon JS. The role of estrogen receptors and their signaling across psychiatric disorders. Int J Mol Sci 2020;22:373.
36. Bernal J. Thyroid hormones in brain development and function. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25905404. [Last accessed on 7 Jun 2021].
38. Lev-Ran S, Roerecke M, Le Foll B, George TP, McKenzie K, Rehm J. The association between cannabis use and depression: a systematic review and meta-analysis of longitudinal studies. Psychol Med 2014;44:797-810.
39. Berger M, Nelson B, Markulev C, et al. Relationship between polyunsaturated fatty acids and psychopathology in the NEURAPRO clinical trial. Front Psychiatry 2019;10:393.
40. Amminger GP, McGorry PD. Update on ω-3 polyunsaturated fatty acids in early-stage psychotic disorders. Neuropsychopharmacology 2012;37:309-10.
41. Olivares AM, Moreno-Ramos OA, Haider NB. Role of nuclear receptors in central nervous system development and associated diseases. J Exp Neurosci 2015;9:93-121.
42. Ferrara SJ, Scanlan TS. A CNS-targeting prodrug strategy for nuclear receptor modulators. J Med Chem 2020;63:9742-51.
43. Mackeh R, Marr AK, Dargham SR, Syed N, Fakhro KA, Kino T. Single-nucleotide variations of the human nuclear hormone receptor genes in 60,000 individuals. J Endocr Soc 2018;2:77-90.
44. Achermann JC, Schwabe J, Fairall L, Chatterjee K. Genetic disorders of nuclear receptors. J Clin Invest 2017;127:1181-92.
45. Guissart C, Latypova X, Rollier P, et al. Dual molecular effects of dominant RORA mutations cause two variants of syndromic intellectual disability with either autism or cerebellar ataxia. Am J Hum Genet 2018;102:744-59.
46. Rudolf G, Lesca G, Mehrjouy MM, et al. Loss of function of the retinoid-related nuclear receptor (RORB) gene and epilepsy. Eur J Hum Genet 2016;24:1761-70.
47. Millard CJ, Watson PJ, Fairall L, Schwabe JW. An evolving understanding of nuclear receptor coregulator proteins. J Mol Endocrinol 2013;51:T23-36.
48. Malovannaya A, Lanz RB, Jung SY, et al. Analysis of the human endogenous coregulator complexome. Cell 2011;145:787-99.
49. Broekema MF, Hollman DAA, Koppen A, et al. Profiling of 3696 nuclear receptor-coregulator interactions: a resource for biological and clinical discovery. Endocrinology 2018;159:2397-407.
50. Tagami T, Madison LD, Nagaya T, Jameson JL. Nuclear receptor corepressors activate rather than suppress basal transcription of genes that are negatively regulated by thyroid hormone. Mol Cell Biol 1997;17:2642-8.
51. Berghagen H, Ragnhildstveit E, Krogsrud K, Thuestad G, Apriletti J, Saatcioglu F. Corepressor SMRT functions as a coactivator for thyroid hormone receptor T3Ralpha from a negative hormone response element. J Biol Chem 2002;277:49517-22.
52. Tora L, Gronemeyer H, Turcotte B, Gaub MP, Chambon P. The N-terminal region of the chicken progesterone receptor specifies target gene activation. Nature 1988;333:185-8.
53. Shao D, Lazar MA. Modulating nuclear receptor function: may the phos be with you. J Clin Invest 1999;103:1617-8.
54. Lonard DM, Lanz RB, O'Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev 2007;28:575-87.
55. Kong Y, Zhou W, Sun Z. Nuclear receptor corepressors in intellectual disability and autism. Mol Psychiatry 2020;25:2220-36.
56. Tabet AC, Leroy C, Dupont C, et al. De novo deletion of TBL1XR1 in a child with non-specific developmental delay supports its implication in intellectual disability. Am J Med Genet A 2014;164A:2335-7.
57. Vaqueiro AC, de Oliveira CP, Cordoba MS, et al. Expanding the spectrum of TBL1XR1 deletion: report of a patient with brain and cardiac malformations. Eur J Med Genet 2018;61:29-33.
58. Sakaguchi Y, Uehara T, Suzuki H, et al. Haploinsufficiency of NCOR1 associated with autism spectrum disorder, scoliosis, and abnormal palatogenesis. Am J Med Genet A 2018;176:2466-9.
59. Zhou W, He Y, Rehman AU, et al. DDD study. Author correction: loss of function of NCOR1 and NCOR2 impairs memory through a novel GABAergic hypothalamus-CA3 projection. Nat Neurosci 2019;22:1533.
60. Sajan SA, Jhangiani SN, Muzny DM, et al. Enrichment of mutations in chromatin regulators in people with Rett syndrome lacking mutations in MECP2. Genet Med 2017;19:13-9.
61. Reay WR, Atkins JR, Quidé Y, Carr VJ, Green MJ, Cairns MJ. Polygenic disruption of retinoid signalling in schizophrenia and a severe cognitive deficit subtype. Mol Psychiatry 2020;25:719-31.
62. Viosca J, Lopez-Atalaya JP, Olivares R, Eckner R, Barco A. Syndromic features and mild cognitive impairment in mice with genetic reduction on p300 activity: differential contribution of p300 and CBP to Rubinstein-Taybi syndrome etiology. Neurobiol Dis 2010;37:186-94.
63. Qvist P, Christensen JH, Vardya I, et al. The schizophrenia-associated BRD1 gene regulates behavior, neurotransmission, and expression of schizophrenia risk enriched gene sets in mice. Biol Psychiatry 2017;82:62-76.
64. Rajkumar AP, Qvist P, Donskov JG, et al. Reduced Brd1 expression leads to reversible depression-like behaviors and gene-expression changes in female mice. Transl Psychiatry 2020;10:239.
65. Qvist P, Eskildsen SF, Hansen B, et al. Brain volumetric alterations accompanied with loss of striatal medium-sized spiny neurons and cortical parvalbumin expressing interneurons in Brd1+/- mice. Sci Rep 2018;8:16486.
66. Oliveira AM, Estévez MA, Hawk JD, Grimes S, Brindle PK, Abel T. Subregion-specific p300 conditional knock-out mice exhibit long-term memory impairments. Learn Mem 2011;18:161-9.
67. Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A. Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation but does not affect cell viability. J Neurosci 2011;31:1652-63.
68. Zhang Z, Hofmann C, Casanova E, Schütz G, Lutz B. Generation of a conditional allele of the CBP gene in mouse. Genesis 2004;40:82-9.
69. Morimoto Y, Ono S, Yoshida S, et al. A unique missense variant in the E1A-binding protein P400 gene is implicated in schizophrenia by whole-exome sequencing and mutant mouse models. Transl Psychiatry 2021;11:132.
70. Ripke S, Walters JTR, O’Donovan MC. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv 2020; doi: 10.1101/2020.09.12.20192922.
71. Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. European Alzheimer's Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer's Disease; Alzheimer's Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013;45:1452-8.
72. Cai L, Wheeler E, Kerrison ND, et al. Genome-wide association analysis of type 2 diabetes in the EPIC-InterAct study. Sci Data 2020;7:393.
73. Shah S, Henry A, Roselli C, et al. Regeneron Genetics Center. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163.
74. Yengo L, Sidorenko J, Kemper KE, et al. GIANT Consortium. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641-9.
75. Leslie R, O'Donnell CJ, Johnson AD. GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics 2014;30:i185-94.
76. Lam M, Awasthi S, Watson HJ, et al. RICOPILI: rapid imputation for COnsortias PIpeLIne. Bioinformatics 2020;36:930-3.
77. Singh T, Kurki MI, Curtis D, et al. Swedish Schizophrenia Study; INTERVAL Study; DDD Study; UK10 K Consortium. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci 2016;19:571-7.
78. Satterstrom FK, Kosmicki JA, Wang J, et al. Autism Sequencing Consortium; iPSYCH-Broad Consortium. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 2020;180:568-84.e23.
79. Hannon E, Dempster E, Viana J, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol 2016;17:176.
80. Aberg KA, McClay JL, Nerella S, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry 2014;71:255-64.
81. Chan RF, Shabalin AA, Montano C, et al. Independent methylome-wide association studies of schizophrenia detect consistent case-control differences. Schizophr Bull 2020;46:319-27.
82. Aberg KA, Dean B, Shabalin AA, et al. Methylome-wide association findings for major depressive disorder overlap in blood and brain and replicate in independent brain samples. Mol Psychiatry 2020;25:1344-54.
83. van Dongen J, Zilhão NR, Sugden K, et al. BIOS Consortium. Epigenome-wide Association Study of Attention-Deficit/Hyperactivity Disorder Symptoms in Adults. Biol Psychiatry 2019;86:599-607.
84. Walton E, Pingault JB, Cecil CA, et al. Epigenetic profiling of ADHD symptoms trajectories: a prospective, methylome-wide study. Mol Psychiatry 2017;22:250-6.
85. Hannon E, Schendel D, Ladd-Acosta C, et al. iPSYCH-Broad ASD Group. Elevated polygenic burden for autism is associated with differential DNA methylation at birth. Genome Med 2018;10:19.
86. Spiers H, Hannon E, Schalkwyk LC, et al. Methylomic trajectories across human fetal brain development. Genome Res 2015;25:338-52.
87. Miller JA, Ding SL, Sunkin SM, et al. Transcriptional landscape of the prenatal human brain. Nature 2014;508:199-206.
88. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015;11:e1004219.
89. Kulakovskiy IV, Vorontsov IE, Yevshin IS, et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res 2018;46:D252-9.
90. Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics 2011;27:1017-8.
91. Shen L, Sinai M. GeneOverlap: Test and visualize gene overlaps. R package version 1.2.0. 2013. Available from: http://shenlab-sinai.github.io/shenlab-sinai/. [Last accessed on 7 Jun 2021].
92. Hoffman GE, Bendl J, Voloudakis G, et al. CommonMind Consortium provides transcriptomic and epigenomic data for Schizophrenia and Bipolar Disorder. Sci Data 2019;6:180.
93. Gearing LJ, Cumming HE, Chapman R, et al. CiiiDER: A tool for predicting and analysing transcription factor binding sites. PLoS One 2019;14:e0215495.
94. Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 2015;43:W566-70.
95. McKenzie AT, Wang M, Hauberg ME, et al. Brain cell type specific gene expression and co-expression network architectures. Sci Rep 2018;8:8868.
96. Shi L, Zhang Z, Su B. Sex biased gene expression profiling of human brains at major developmental stages. Sci Rep 2016;6:21181.
97. Buroker NE, Young ME, Wei C, et al. The dominant negative thyroid hormone receptor beta-mutant {Delta}337T alters PPAR{alpha} signaling in heart. Am J Physiol Endocrinol Metab 2007;292:E453-60.
98. Swanson HI, Wada T, Xie W, et al. Role of nuclear receptors in lipid dysfunction and obesity-related diseases. Drug Metab Dispos 2013;41:1-11.
99. Liu S, Downes M, Evans RM. Metabolic regulation by nuclear receptors. In: Nakao K, Minato N, Uemoto S, editors. Innovative medicine. Tokyo: Springer Japan; 2015. p. 25-37.
100. Moutinho M, Landreth GE. Therapeutic potential of nuclear receptor agonists in Alzheimer's disease. J Lipid Res 2017;58:1937-49.
101. Fitz NF, Nam KN, Koldamova R, Lefterov I. Therapeutic targeting of nuclear receptors, liver X and retinoid X receptors, for Alzheimer's disease. Br J Pharmacol 2019;176:3599-610.
102. Corredor B, Dattani M, Gertosio C, Bozzola M. Tall stature: a challenge for clinicians. Curr Pediatr Rev 2019;15:10-21.
103. De Magalhaes Filho CD, Downes M, Evans RM. Farnesoid X receptor an emerging target to combat obesity. Dig Dis 2017;35:185-90.
104. Skerrett R, Malm T, Landreth G. Nuclear receptors in neurodegenerative diseases. Neurobiol Dis 2014;72 Pt A:104-16.
105. Rees E, Owen MJ. Translating insights from neuropsychiatric genetics and genomics for precision psychiatry. Genome Med 2020;12:43.
106. Owen MJ, O'Donovan MC. Schizophrenia and the neurodevelopmental continuum:evidence from genomics. World Psychiatry 2017;16:227-35.
107. Aberg KA, Xie LY, McClay JL, et al. Testing two models describing how methylome-wide studies in blood are informative for psychiatric conditions. Epigenomics 2013;5:367-77.
108. Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 2008;82:696-711.
109. Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry 2014;19:862-71.
110. Starnawska A, Tan Q, McGue M, et al. Epigenome-wide association study of cognitive functioning in middle-aged monozygotic twins. Front Aging Neurosci 2017;9:413.
111. Ianov L, Riva A, Kumar A, Foster TC. DNA methylation of synaptic genes in the prefrontal cortex is associated with aging and age-related cognitive impairment. Front Aging Neurosci 2017;9:249.
112. Vogel Ciernia A, LaSalle J. The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nat Rev Neurosci 2016;17:411-23.
113. Montano C, Taub MA, Jaffe A, et al. Association of DNA methylation differences with schizophrenia in an epigenome-wide association study. JAMA Psychiatry 2016;73:506-14.
114. Córdova-Palomera A, Fatjó-Vilas M, Gastó C, Navarro V, Krebs MO, Fañanás L. Genome-wide methylation study on depression: differential methylation and variable methylation in monozygotic twins. Transl Psychiatry 2015;5:e557.
115. Palma-Gudiel H, Córdova-Palomera A, Eixarch E, Deuschle M, Fañanás L. Maternal psychosocial stress during pregnancy alters the epigenetic signature of the glucocorticoid receptor gene promoter in their offspring: a meta-analysis. Epigenetics 2015;10:893-902.
116. Chen S, Mukherjee N, Janjanam VD, et al. Consistency and variability of DNA methylation in women during puberty, young adulthood, and pregnancy. Genet Epigenet 2017;9:1179237X17721540.
117. Lomniczi A, Loche A, Castellano JM, et al. Epigenetic control of female puberty. Nat Neurosci 2013;16:281-9.
119. Corley SM, Tsai SY, Wilkins MR, Shannon Weickert C. Transcriptomic analysis shows decreased cortical expression of NR4A1, NR4A2 and RXRB in schizophrenia and provides evidence for nuclear receptor dysregulation. PLoS One 2016;11:e0166944.
120. Maire A, Teyssier C, Balaguer P, Bourguet W, Germain P. Regulation of RXR-RAR heterodimers by RXR- and RAR-specific ligands and their combinations. Cells 2019;8:1392.
121. Penvose A, Keenan JL, Bray D, Ramlall V, Siggers T. Comprehensive study of nuclear receptor DNA binding provides a revised framework for understanding receptor specificity. Nat Commun 2019;10:2514.
122. Lenroot RK, Gogtay N, Greenstein DK, et al. Sexual dimorphism of brain developmental trajectories during childhood and adolescence. Neuroimage 2007;36:1065-73.
123. Marrocco J, McEwen BS. Sex in the brain: hormones and sex differences. Dialogues Clin Neurosci 2017;18:373-83.
124. Hu VW, Sarachana T, Sherrard RM, Kocher KM. Investigation of sex differences in the expression of RORA and its transcriptional targets in the brain as a potential contributor to the sex bias in autism. Mol Autism 2015;6:7.
125. Lenroot RK, Giedd JN. The changing impact of genes and environment on brain development during childhood and adolescence: initial findings from a neuroimaging study of pediatric twins. Dev Psychopathol 2008;20:1161-75.
126. Ahmed OM, El-Gareib AW, El-Bakry AM, Abd El-Tawab SM, Ahmed RG. Thyroid hormones states and brain development interactions. Int J Dev Neurosci 2008;26:147-209.
127. Herzmann C, Torrens JK. Maternal thyroid deficiency during pregnancy and subsequent neuropsychological development of the child. N Engl J Med 1999;341:2015.
128. Zoeller RT, Rovet J. Timing of thyroid hormone action in the developing brain: clinical observations and experimental findings. J Neuroendocrinol 2004;16:809-18.
129. Pertile RA, Cui X, Eyles DW. Vitamin D signaling and the differentiation of developing dopamine systems. Neuroscience 2016;333:193-203.
130. Labrie F, Bélanger A, Cusan L, Gomez JL, Candas B. Marked decline in serum concentrations of adrenal C19 sex steroid precursors and conjugated androgen metabolites during aging. J Clin Endocrinol Metab 1997;82:2396-402.
131. Baron-Cohen S, Auyeung B, Nørgaard-Pedersen B, et al. Elevated fetal steroidogenic activity in autism. Mol Psychiatry 2015;20:369-76.
132. Holsboer F. The corticosteroid receptor hypothesis of depression. Neuropsychopharmacology 2000;23:477-501.
133. Xu D, Huang S, Wang H, Xie W. Regulation of brain drug metabolizing enzymes and transporters by nuclear receptors. Drug Metab Rev 2018;50:407-14.
134. Erk S, Mohnke S, Ripke S, et al. Functional neuroimaging effects of recently discovered genetic risk loci for schizophrenia and polygenic risk profile in five RDoC subdomains. Transl Psychiatry 2017;7:e997.
135. Hukkanen J, Hakkola J, Rysä J. Pregnane X receptor (PXR)--a contributor to the diabetes epidemic? Drug Metabol Drug Interact 2014;29:3-15.
136. Gao J, He J, Zhai Y, Wada T, Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J Biol Chem 2009;284:25984-92.
137. Cipolletta D, Feuerer M, Li A, et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 2012;486:549-53.
138. Dalgård C, Petersen MS, Weihe P, Grandjean P. Vitamin D status in relation to glucose metabolism and type 2 diabetes in septuagenarians. Diabetes Care 2011;34:1284-8.
139. Cummings JL, Zhong K, Kinney JW, et al. Double-blind, placebo-controlled, proof-of-concept trial of bexarotene Xin moderate Alzheimer's disease. Alzheimers Res Ther 2016;8:4.
140. Gordon JL, Peltier A, Grummisch JA, Sykes Tottenham L. Estradiol fluctuation, sensitivity to stress, and depressive symptoms in the menopause transition: a pilot study. Front Psychol 2019;10:1319.
141. Thomas N, Gurvich C, Hudaib AR, Gavrilidis E, Kulkarni J. Dissecting the syndrome of schizophrenia: associations between symptomatology and hormone levels in women with schizophrenia. Psychiatry Res 2019;280:112510.
142. Dwyer JB, Aftab A, Radhakrishnan R, et al. APA Council of Research Task Force on Novel Biomarkers and Treatments. Hormonal treatments for major depressive disorder: state of the art. Am J Psychiatry 2020;177:686-705.
143. Meinhard N, Kessing LV, Vinberg M. The role of estrogen in bipolar disorder, a review. Nord J Psychiatry 2014;68:81-7.
144. de Boer J, Prikken M, Lei WU, Begemann M, Sommer I. The effect of raloxifene augmentation in men and women with a schizophrenia spectrum disorder: a systematic review and meta-analysis. NPJ Schizophr 2018;4:1.
145. Popovic D, Schmitt A, Kaurani L, et al. Childhood trauma in schizophrenia: current findings and research perspectives. Front Neurosci 2019;13:274.
146. Aas M, Henry C, Andreassen OA, Bellivier F, Melle I, Etain B. The role of childhood trauma in bipolar disorders. Int J Bipolar Disord 2016;4:2.
147. ter Heegde F, De Rijk RH, Vinkers CH. The brain mineralocorticoid receptor and stress resilience. Psychoneuroendocrinology 2015;52:92-110.
148. Endedijk HM, Nelemans SA, Schür RR, et al. The role of stress and mineralocorticoid receptor haplotypes in the development of symptoms of depression and anxiety during adolescence. Front Psychiatry 2020;11:367.
149. Vinkers CH, Joëls M, Milaneschi Y, et al. Mineralocorticoid receptor haplotypes sex-dependently moderate depression susceptibility following childhood maltreatment. Psychoneuroendocrinology 2015;54:90-102.
150. Bordoni A, Di Nunzio M, Danesi F, Biagi PL. Polyunsaturated fatty acids: from diet to binding to ppars and other nuclear receptors. Genes Nutr 2006;1:95-106.
151. Lengqvist J, Mata De Urquiza A, Bergman AC, et al. Polyunsaturated fatty acids including docosahexaenoic and arachidonic acid bind to the retinoid X receptor alpha ligand-binding domain. Mol Cell Proteomics 2004;3:692-703.
152. van Neerven S, Kampmann E, Mey J. RAR/RXR and PPAR/RXR signaling in neurological and psychiatric diseases. Prog Neurobiol 2008;85:433-51.
153. O'Sullivan SE. Cannabinoids go nuclear: evidence for activation of peroxisome proliferator-activated receptors. Br J Pharmacol 2007;152:576-82.
154. Leweke FM, Koethe D. Cannabis and psychiatric disorders: it is not only addiction. Addict Biol 2008;13:264-75.
155. Velmeshev D, Schirmer L, Jung D, et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science 2019;364:685-9.
156. Sarachana T, Hu VW. Genome-wide identification of transcriptional targets of RORA reveals direct regulation of multiple genes associated with autism spectrum disorder. Mol Autism 2013;4:14.
157. Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov 2014;13:197-216.
158. Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J 2010;24:3036-51.
159. Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal 2009;7:e003.
160. Grayaa S, Zerbinati C, Messedi M, et al. Plasma oxysterol profiling in children reveals 24-hydroxycholesterol as a potential marker for Autism Spectrum Disorders. Biochimie 2018;153:80-5.
161. de Vera IM, Giri PK, Munoz-Tello P, et al. Identification of a binding site for unsaturated fatty acids in the orphan nuclear receptor nurr1. ACS Chem Biol 2016;11:1795-9.
162. Willems S, Kilu W, Ni X, et al. The orphan nuclear receptor Nurr1 is responsive to non-steroidal anti-inflammatory drugs. Commun Chem 2020;3:85.
163. Pan T, Zhu W, Zhao H, et al. Nurr1 deficiency predisposes to lactacystin-induced dopaminergic neuron injury in vitro and in vivo. Brain Res 2008;1222:222-9.
164. Decressac M, Volakakis N, Björklund A, Perlmann T. NURR1 in Parkinson disease--from pathogenesis to therapeutic potential. Nat Rev Neurol 2013;9:629-36.
165. Zhou J, Oakley RH, Cidlowski JA. DAX-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X-chromosome, gene 1) selectively inhibits transactivation but not transrepression mediated by the glucocorticoid receptor in a LXXLL-dependent manner. Mol Endocrinol 2008;22:1521-34.
166. Obradović D, Tirard M, Némethy Z, Hirsch O, Gronemeyer H, Almeida OF. DAXX, FLASH, and FAF-1 modulate mineralocorticoid and glucocorticoid receptor-mediated transcription in hippocampal cells--toward a basis for the opposite actions elicited by two nuclear receptors? Mol Pharmacol 2004;65:761-9.
167. Zhao L, Zhou S, Gustafsson JÅ. Nuclear receptors: recent drug discovery for cancer therapies. Endocr Rev 2019;40:1207-49.
168. Santos R, Ursu O, Gaulton A, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov 2017;16:19-34.
169. Marciano DP, Chang MR, Corzo CA, et al. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARγ, RORs, and Rev-erbs. Cell Metab 2014;19:193-208.
170. McKenna NJ. Research resources for nuclear receptor signaling pathways. Mol Pharmacol 2016;90:153-9.
171. Corona JC, Duchen MR. PPARγ as a therapeutic target to rescue mitochondrial function in neurological disease. Free Radic Biol Med 2016;100:153-63.
172. Moutinho M, Codocedo JF, Puntambekar SS, Landreth GE. Nuclear receptors as therapeutic targets for neurodegenerative diseases: lost in translation. Annu Rev Pharmacol Toxicol 2019;59:237-61.
173. Kim CH, Han BS, Moon J, et al. Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson's disease. Proc Natl Acad Sci U S A 2015;112:8756-61.
174. Tong M, Dominguez C, Didsbury J, de la Monte SM. Targeting Alzheimer's disease neuro-metabolic dysfunction with a small molecule nuclear receptor agonist (T3D-959) reverses disease pathologies. J Alzheimers Dis Parkinsonism 2016;6:238.
175. Sharma RP. Schizophrenia, epigenetics and ligand-activated nuclear receptors: a framework for chromatin therapeutics. Schizophr Res 2005;72:79-90.
176. Carta MG, Paribello P, Preti A. How promising is neuroactive steroid drug discovery? Expert Opin Drug Discov 2018;13:993-5.
177. Lerner V, Miodownik C, Gibel A, et al. The retinoid X receptor agonist bexarotene relieves positive symptoms of schizophrenia: a 6-week, randomized, double-blind, placebo-controlled multicenter trial. J Clin Psychiatry 2013;74:1224-32.
178. Touma KTB, Zoucha AM, Scarff JR. Liothyronine for depression: a review and guidance for safety monitoring. Innov Clin Neurosci 2017;14:24-9.
180. Song X, Chen J, Zhao M, et al. Development of potent small-molecule inhibitors to drug the undruggable steroid receptor coactivator-3. Proc Natl Acad Sci U S A 2016;113:4970-5.
181. Raj GV, Sareddy GR, Ma S, et al. Estrogen receptor coregulator binding modulators (ERXs) effectively target estrogen receptor positive human breast cancers. Elife 2017;6:e26857.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Donskov JG, Starnawska A, Pallesen J, Grove J, Børglum AD, Qvist P. Large-scale genomic data-mining implicates dysregulated nuclear receptor-mediated signaling in mental illness. J Transl Genet Genom 2021;5:136-62. http://dx.doi.org/10.20517/jtgg.2021.12
AMA Style
Donskov JG, Starnawska A, Pallesen J, Grove J, Børglum AD, Qvist P. Large-scale genomic data-mining implicates dysregulated nuclear receptor-mediated signaling in mental illness. Journal of Translational Genetics and Genomics. 2021; 5(2): 136-62. http://dx.doi.org/10.20517/jtgg.2021.12
Chicago/Turabian Style
Donskov, Julie G., Anna Starnawska, Jonatan Pallesen, Jakob Grove, Anders D. Børglum, Per Qvist. 2021. "Large-scale genomic data-mining implicates dysregulated nuclear receptor-mediated signaling in mental illness" Journal of Translational Genetics and Genomics. 5, no.2: 136-62. http://dx.doi.org/10.20517/jtgg.2021.12
ACS Style
Donskov, JG.; Starnawska A.; Pallesen J.; Grove J.; Børglum AD.; Qvist P. Large-scale genomic data-mining implicates dysregulated nuclear receptor-mediated signaling in mental illness. J. Transl. Genet. Genom. 2021, 5, 136-62. http://dx.doi.org/10.20517/jtgg.2021.12
About This Article
Special Issue
Copyright
Data & Comments
Data

 Cite This Article 37 clicks
Cite This Article 37 clicks


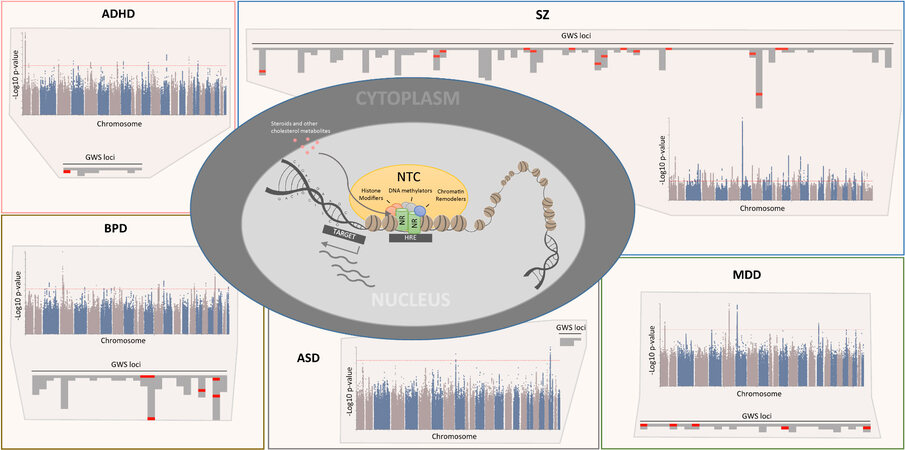
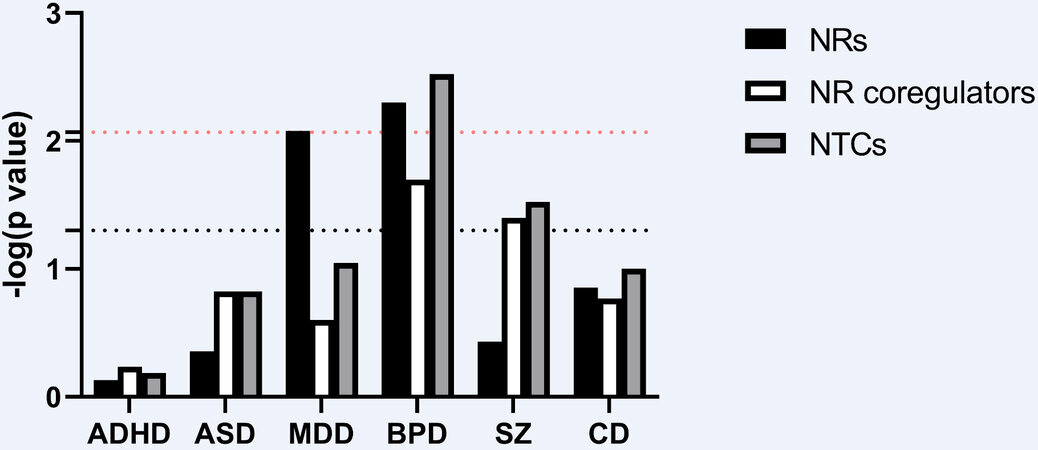
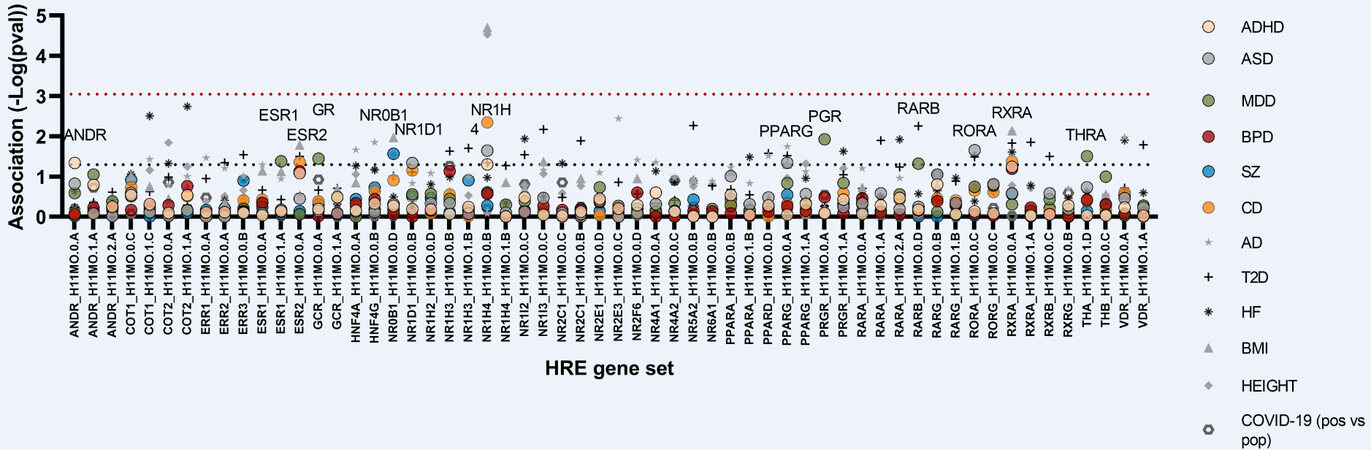
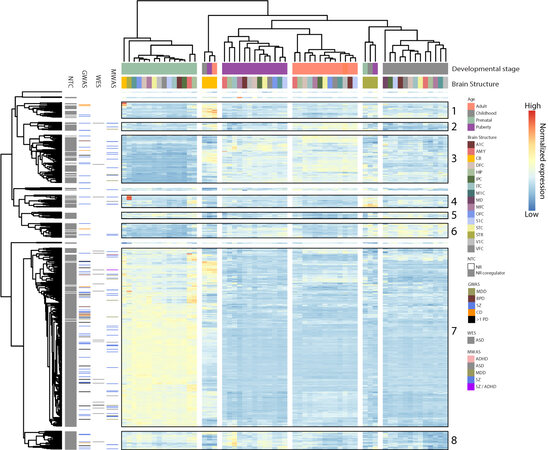
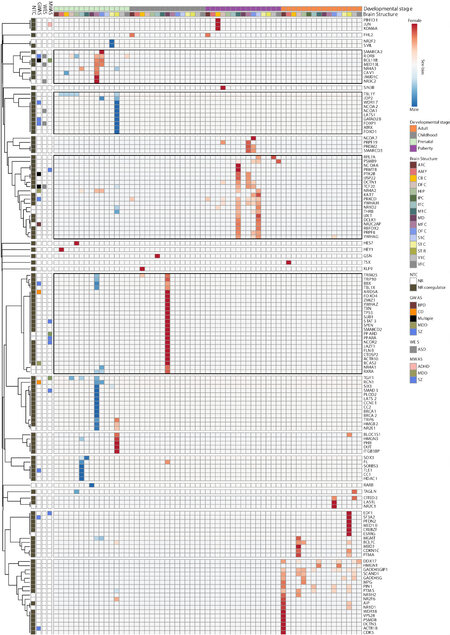
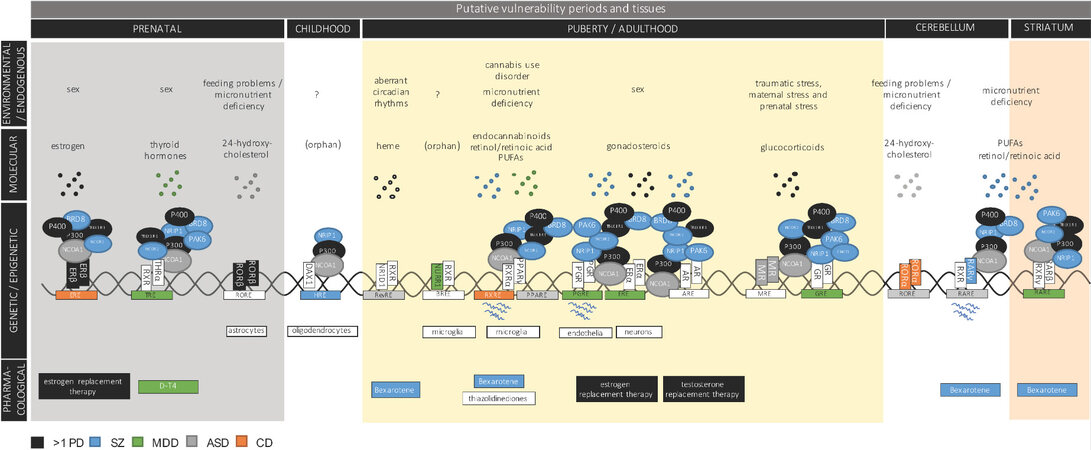








Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.